The Alternative Sigma Factor RpoE2 Is Involved in the Stress Response to Hypochlorite and in vivo Survival of Haemophilus influenzae
- PMID: 33643271
- PMCID: PMC7907618
- DOI: 10.3389/fmicb.2021.637213
The Alternative Sigma Factor RpoE2 Is Involved in the Stress Response to Hypochlorite and in vivo Survival of Haemophilus influenzae
Abstract
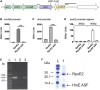
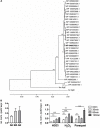
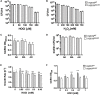
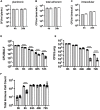
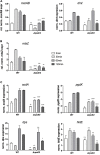
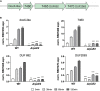
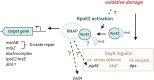
Extracytoplasmic function (ECF) sigma factors underpin the ability of bacteria to adapt to changing environmental conditions, a process that is particularly relevant in human pathogens that inhabit niches where human immune cells contribute to high levels of extracellular stress. Here, we have characterized the previously unstudied RpoE2 ECF sigma factor from the human respiratory pathogen H. influenzae (Hi) and its role in hypochlorite-induced stress. Exposure of H. influenzae to oxidative stress (HOCl, H2O2) increased rpoE2 gene expression, and the activity of RpoE2 was controlled by a cytoplasmic 67-aa anti-sigma factor, HrsE. RpoE2 regulated the expression of the periplasmic MsrAB peptide methionine sulfoxide reductase that, in H. influenzae, is required for HOCl resistance, thus linking RpoE2 to HOCl stress. Interestingly, a HiΔrpoE2 strain had wild-type levels of resistance to oxidative stress in vitro, but HiΔrpoE2 survival was reduced 26-fold in a mouse model of lung infection, demonstrating the relevance of this sigma factor for H. influenzae pathogenesis. The HiRpoE2 system has some similarity to the ECF sigma factors described in Streptomyces and Neisseria sp. that also control the expression of msr genes. However, HiRpoE2 regulation extended to genes encoding other periplasmic damage repair proteins, an operon containing a DoxX-like protein, and also included selected OxyR-controlled genes. Based on our results, we propose that the highly conserved HiRpoE2 sigma factor is a key regulator of H. influenzae responses to oxidative damage in the cell envelope region that controls a variety of target genes required for survival in the host.
Keywords: H. influenzae; extracytoplasmic function sigma factor; gene regulation; hypochlorite; stress response.
Copyright © 2021 Nasreen, Fletcher, Hosmer, Zhong, Essilfie, McEwan and Kappler.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
An extracytoplasmic function sigma factor-dependent periplasmic glutathione peroxidase is involved in oxidative stress response of Shewanella oneidensis.BMC Microbiol. 2015 Feb 18;15:34. doi: 10.1186/s12866-015-0357-0. BMC Microbiol. 2015. PMID: 25887418 Free PMC article.
-
Peptide Methionine Sulfoxide Reductase from Haemophilus influenzae Is Required for Protection against HOCl and Affects the Host Response to Infection.ACS Infect Dis. 2020 Jul 10;6(7):1928-1939. doi: 10.1021/acsinfecdis.0c00242. Epub 2020 Jun 22. ACS Infect Dis. 2020. PMID: 32492342
-
An extracytoplasmic function sigma factor acts as a general stress response regulator in Sinorhizobium meliloti.J Bacteriol. 2007 Jun;189(11):4204-16. doi: 10.1128/JB.00175-07. Epub 2007 Mar 30. J Bacteriol. 2007. PMID: 17400745 Free PMC article.
-
Diversity of extracytoplasmic function sigma (σECF ) factor-dependent signaling in Pseudomonas.Mol Microbiol. 2019 Aug;112(2):356-373. doi: 10.1111/mmi.14331. Epub 2019 Jul 3. Mol Microbiol. 2019. PMID: 31206859 Review.
-
New insights into the molecular physiology of sulfoxide reduction in bacteria.Adv Microb Physiol. 2019;75:1-51. doi: 10.1016/bs.ampbs.2019.05.001. Epub 2019 Jul 5. Adv Microb Physiol. 2019. PMID: 31655735 Review.
Cited by
-
The Peptide Methionine Sulfoxide Reductase (MsrAB) of Haemophilus influenzae Repairs Oxidatively Damaged Outer Membrane and Periplasmic Proteins Involved in Nutrient Acquisition and Virulence.Antioxidants (Basel). 2022 Aug 11;11(8):1557. doi: 10.3390/antiox11081557. Antioxidants (Basel). 2022. PMID: 36009276 Free PMC article.
-
A multi-iron enzyme installs copper-binding oxazolone/thioamide pairs on a nontypeable Haemophilus influenzae virulence factor.Proc Natl Acad Sci U S A. 2024 Jul 9;121(28):e2408092121. doi: 10.1073/pnas.2408092121. Epub 2024 Jul 5. Proc Natl Acad Sci U S A. 2024. PMID: 38968106 Free PMC article.
-
The DmsABC S-oxide reductase is an essential component of a novel, hypochlorite-inducible system of extracellular stress defense in Haemophilus influenzae.Front Microbiol. 2024 Apr 4;15:1359513. doi: 10.3389/fmicb.2024.1359513. eCollection 2024. Front Microbiol. 2024. PMID: 38638903 Free PMC article.
-
Hfe Permease and Haemophilus influenzae Manganese Homeostasis.ACS Infect Dis. 2024 Feb 9;10(2):436-452. doi: 10.1021/acsinfecdis.3c00407. Epub 2024 Jan 19. ACS Infect Dis. 2024. PMID: 38240689 Free PMC article.
References
-
- Ausubel F. M. (2002). Short Protocols in Molecular Biology: A Compendium of Methods from “Current Protocols in Molecular Biology”. New York, NY: Wiley.
-
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., et al. (2005). “Current protocols in molecular biology,” in Current Protocols in Molecular Biology, ed. Janssen K. (Hoboken, NJ: John Wiley & Sons Inc; ).
-
- Coligan J. E. (2003). Short Protocols in Protein Science: A Compendium of Methods from Current Protocols in Protein Science. Hoboken, N.J: Wiley.
LinkOut - more resources
Full Text Sources
Other Literature Sources

