Specific PIP2 binding promotes calcium activation of TMEM16A chloride channels
- PMID: 33637964
- PMCID: PMC7910439
- DOI: 10.1038/s42003-021-01782-2
Specific PIP2 binding promotes calcium activation of TMEM16A chloride channels
Abstract
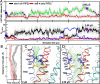
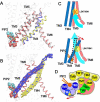
TMEM16A is a widely expressed Ca2+-activated Cl- channel that regulates crucial physiological functions including fluid secretion, neuronal excitability, and smooth muscle contraction. There is a critical need to understand the molecular mechanisms of TMEM16A gating and regulation. However, high-resolution TMEM16A structures have failed to reveal an activated state with an unobstructed permeation pathway even with saturating Ca2+. This has been attributed to the requirement of PIP2 for preventing TMEM16A desensitization. Here, atomistic simulations show that specific binding of PIP2 to TMEM16A can lead to spontaneous opening of the permeation pathway in the Ca2+-bound state. The predicted activated state is highly consistent with a wide range of mutagenesis and functional data. It yields a maximal Cl- conductance of ~1 pS, similar to experimental estimates, and recapitulates the selectivity of larger SCN- over Cl-. The resulting molecular mechanism of activation provides a basis for understanding the interplay of multiple signals in controlling TMEM16A channel function.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Molecular basis of PIP2-dependent regulation of the Ca2+-activated chloride channel TMEM16A.Nat Commun. 2019 Aug 21;10(1):3769. doi: 10.1038/s41467-019-11784-8. Nat Commun. 2019. PMID: 31434906 Free PMC article.
-
Revealing the activation pathway for TMEM16A chloride channels from macroscopic currents and kinetic models.Pflugers Arch. 2016 Jul;468(7):1241-1257. doi: 10.1007/s00424-016-1830-9. Epub 2016 May 2. Pflugers Arch. 2016. PMID: 27138167 Free PMC article.
-
Phosphatidylinositol 4,5-bisphosphate (PIP2) and Ca2+ are both required to open the Cl- channel TMEM16A.J Biol Chem. 2019 Aug 16;294(33):12556-12564. doi: 10.1074/jbc.RA118.007128. Epub 2019 Jul 2. J Biol Chem. 2019. PMID: 31266809 Free PMC article.
-
Wasted TMEM16A channels are rescued by phosphatidylinositol 4,5-bisphosphate.Cell Calcium. 2019 Dec;84:102103. doi: 10.1016/j.ceca.2019.102103. Epub 2019 Oct 18. Cell Calcium. 2019. PMID: 31683182 Free PMC article. Review.
-
Insights into the function and regulation of the calcium-activated chloride channel TMEM16A.Cell Calcium. 2024 Jul;121:102891. doi: 10.1016/j.ceca.2024.102891. Epub 2024 May 8. Cell Calcium. 2024. PMID: 38772195 Review.
Cited by
-
Simulation-based survey of TMEM16 family reveals that robust lipid scrambling requires an open groove.bioRxiv [Preprint]. 2024 Nov 13:2024.09.25.615027. doi: 10.1101/2024.09.25.615027. bioRxiv. 2024. PMID: 39386458 Free PMC article. Preprint.
-
Structure and Function of Calcium-Activated Chloride Channels and Phospholipid Scramblases in the TMEM16 Family.Handb Exp Pharmacol. 2024;283:153-180. doi: 10.1007/164_2022_595. Handb Exp Pharmacol. 2024. PMID: 35792944 Review.
-
Activation of TMEM16F by inner gate charged mutations and possible lipid/ion permeation mechanisms.Biophys J. 2022 Sep 20;121(18):3445-3457. doi: 10.1016/j.bpj.2022.08.011. Epub 2022 Aug 17. Biophys J. 2022. PMID: 35978550 Free PMC article.
-
Inhibition mechanism of the chloride channel TMEM16A by the pore blocker 1PBC.Nat Commun. 2022 May 19;13(1):2798. doi: 10.1038/s41467-022-30479-1. Nat Commun. 2022. PMID: 35589730 Free PMC article.
-
Niclosamide potentiates TMEM16A and induces vasoconstriction.J Gen Physiol. 2024 Jul 1;156(7):e202313460. doi: 10.1085/jgp.202313460. Epub 2024 May 30. J Gen Physiol. 2024. PMID: 38814250 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

