Cytoplasmic cleavage of IMPA1 3' UTR is necessary for maintaining axon integrity
- PMID: 33626357
- PMCID: PMC7918530
- DOI: 10.1016/j.celrep.2021.108778
Cytoplasmic cleavage of IMPA1 3' UTR is necessary for maintaining axon integrity
Abstract
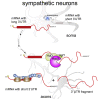
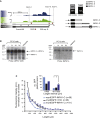
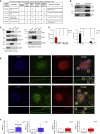
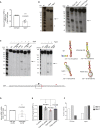
The 3' untranslated regions (3' UTRs) of messenger RNAs (mRNAs) are non-coding sequences involved in many aspects of mRNA metabolism, including intracellular localization and translation. Incorrect processing and delivery of mRNA cause severe developmental defects and have been implicated in many neurological disorders. Here, we use deep sequencing to show that in sympathetic neuron axons, the 3' UTRs of many transcripts undergo cleavage, generating isoforms that express the coding sequence with a short 3' UTR and stable 3' UTR-derived fragments of unknown function. Cleavage of the long 3' UTR of Inositol Monophosphatase 1 (IMPA1) mediated by a protein complex containing the endonuclease argonaute 2 (Ago2) generates a translatable isoform that is necessary for maintaining the integrity of sympathetic neuron axons. Thus, our study provides a mechanism of mRNA metabolism that simultaneously regulates local protein synthesis and generates an additional class of 3' UTR-derived RNAs.
Keywords: 3’UTR; 3’UTR cleavage; NGF; RNA processing; alternative polyadenylation; axons; local translation; mRNA localization; neuronal development; sympathetic neurons.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


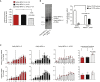
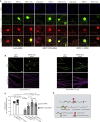
Similar articles
-
An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons.Nat Neurosci. 2010 Mar;13(3):291-301. doi: 10.1038/nn.2486. Epub 2010 Jan 31. Nat Neurosci. 2010. PMID: 20118926
-
A detailed protocol for RNA cleavage assay in sympathetic neurons.STAR Protoc. 2021 Dec 11;2(4):101001. doi: 10.1016/j.xpro.2021.101001. eCollection 2021 Dec 17. STAR Protoc. 2021. PMID: 34950884 Free PMC article.
-
Axonal localization of neuritin/CPG15 mRNA is limited by competition for HuD binding.J Cell Sci. 2017 Nov 1;130(21):3650-3662. doi: 10.1242/jcs.201244. Epub 2017 Sep 4. J Cell Sci. 2017. PMID: 28871047 Free PMC article.
-
Emerging Roles for 3' UTRs in Neurons.Int J Mol Sci. 2020 May 12;21(10):3413. doi: 10.3390/ijms21103413. Int J Mol Sci. 2020. PMID: 32408514 Free PMC article. Review.
-
Implications of polyadenylation in health and disease.Nucleus. 2014;5(6):508-19. doi: 10.4161/nucl.36360. Epub 2014 Oct 31. Nucleus. 2014. PMID: 25484187 Free PMC article. Review.
Cited by
-
Alternative Transcripts Diversify Genome Function for Phenome Relevance to Health and Diseases.Genes (Basel). 2023 Nov 8;14(11):2051. doi: 10.3390/genes14112051. Genes (Basel). 2023. PMID: 38002994 Free PMC article. Review.
-
Injury primes mutation-bearing astrocytes for dedifferentiation in later life.Curr Biol. 2023 Mar 27;33(6):1082-1098.e8. doi: 10.1016/j.cub.2023.02.013. Epub 2023 Feb 24. Curr Biol. 2023. PMID: 36841240 Free PMC article.
-
The predicted RNA-binding protein regulome of axonal mRNAs.Genome Res. 2023 Sep;33(9):1497-1512. doi: 10.1101/gr.277804.123. Epub 2023 Aug 15. Genome Res. 2023. PMID: 37582635 Free PMC article.
-
Regulation and function of alternative polyadenylation in development and differentiation.RNA Biol. 2023 Jan;20(1):908-925. doi: 10.1080/15476286.2023.2275109. Epub 2023 Oct 31. RNA Biol. 2023. PMID: 37906624 Free PMC article. Review.
-
Widespread 3'UTR capped RNAs derive from G-rich regions in proximity to AGO2 binding sites.BMC Biol. 2024 Nov 7;22(1):254. doi: 10.1186/s12915-024-02032-7. BMC Biol. 2024. PMID: 39511645 Free PMC article.
References
-
- Aakalu G., Smith W.B., Nguyen N., Jiang C., Schuman E.M. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. - PubMed
-
- Andreassi C., Zimmermann C., Mitter R., Fusco S., De Vita S., Saiardi A., Riccio A. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat. Neurosci. 2010;13:291–301. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

