Full structural ensembles of intrinsically disordered proteins from unbiased molecular dynamics simulations
- PMID: 33623120
- PMCID: PMC7902620
- DOI: 10.1038/s42003-021-01759-1
Full structural ensembles of intrinsically disordered proteins from unbiased molecular dynamics simulations
Abstract
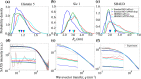
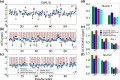
Molecular dynamics (MD) simulation is widely used to complement ensemble-averaged experiments of intrinsically disordered proteins (IDPs). However, MD often suffers from limitations of inaccuracy. Here, we show that enhancing the sampling using Hamiltonian replica-exchange MD (HREMD) led to unbiased and accurate ensembles, reproducing small-angle scattering and NMR chemical shift experiments, for three IDPs of varying sequence properties using two recently optimized force fields, indicating the general applicability of HREMD for IDPs. We further demonstrate that, unlike HREMD, standard MD can reproduce experimental NMR chemical shifts, but not small-angle scattering data, suggesting chemical shifts are insufficient for testing the validity of IDP ensembles. Surprisingly, we reveal that despite differences in their sequence, the inter-chain statistics of all three IDPs are similar for short contour lengths (< 10 residues). The results suggest that the major hurdle of generating an accurate unbiased ensemble for IDPs has now been largely overcome.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Molecular Dynamics Simulations Combined with Nuclear Magnetic Resonance and/or Small-Angle X-ray Scattering Data for Characterizing Intrinsically Disordered Protein Conformational Ensembles.J Chem Inf Model. 2019 May 28;59(5):1743-1758. doi: 10.1021/acs.jcim.8b00928. Epub 2019 Mar 18. J Chem Inf Model. 2019. PMID: 30840442 Review.
-
Structural characterization of an intrinsically disordered protein complex using integrated small-angle neutron scattering and computing.Protein Sci. 2023 Oct;32(10):e4772. doi: 10.1002/pro.4772. Protein Sci. 2023. PMID: 37646172 Free PMC article.
-
Generation of the configurational ensemble of an intrinsically disordered protein from unbiased molecular dynamics simulation.Proc Natl Acad Sci U S A. 2019 Oct 8;116(41):20446-20452. doi: 10.1073/pnas.1907251116. Epub 2019 Sep 23. Proc Natl Acad Sci U S A. 2019. PMID: 31548393 Free PMC article.
-
Development of Charge-Augmented Three-Point Water Model (CAIPi3P) for Accurate Simulations of Intrinsically Disordered Proteins.Int J Mol Sci. 2020 Aug 26;21(17):6166. doi: 10.3390/ijms21176166. Int J Mol Sci. 2020. PMID: 32859072 Free PMC article.
-
Recent Advances in Computational Protocols Addressing Intrinsically Disordered Proteins.Biomolecules. 2019 Apr 11;9(4):146. doi: 10.3390/biom9040146. Biomolecules. 2019. PMID: 30979035 Free PMC article. Review.
Cited by
-
Intrinsically disordered region amplifies membrane remodeling to augment selective ER-phagy.Proc Natl Acad Sci U S A. 2024 Oct 29;121(44):e2408071121. doi: 10.1073/pnas.2408071121. Epub 2024 Oct 25. Proc Natl Acad Sci U S A. 2024. PMID: 39453744
-
Advanced Sampling Methods for Multiscale Simulation of Disordered Proteins and Dynamic Interactions.Biomolecules. 2021 Sep 28;11(10):1416. doi: 10.3390/biom11101416. Biomolecules. 2021. PMID: 34680048 Free PMC article. Review.
-
Clustering Heterogeneous Conformational Ensembles of Intrinsically Disordered Proteins with t-Distributed Stochastic Neighbor Embedding.J Chem Theory Comput. 2023 Jul 25;19(14):4711-4727. doi: 10.1021/acs.jctc.3c00224. Epub 2023 Jun 20. J Chem Theory Comput. 2023. PMID: 37338049 Free PMC article.
-
Self-Diffusive Properties of the Intrinsically Disordered Protein Histatin 5 and the Impact of Crowding Thereon: A Combined Neutron Spectroscopy and Molecular Dynamics Simulation Study.J Phys Chem B. 2022 Feb 3;126(4):789-801. doi: 10.1021/acs.jpcb.1c08976. Epub 2022 Jan 19. J Phys Chem B. 2022. PMID: 35044776 Free PMC article.
-
The diversity of molecular interactions involving intrinsically disordered proteins: A molecular modeling perspective.Comput Struct Biotechnol J. 2021 Jun 24;19:3817-3828. doi: 10.1016/j.csbj.2021.06.031. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34285781 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

