Positive feedback of SuFu negating protein 1 on Hedgehog signaling promotes colorectal tumor growth
- PMID: 33608498
- PMCID: PMC7896051
- DOI: 10.1038/s41419-021-03487-0
Positive feedback of SuFu negating protein 1 on Hedgehog signaling promotes colorectal tumor growth
Abstract
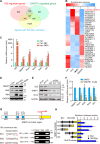
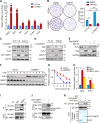
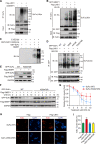
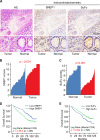
Hedgehog (Hh) signaling plays a critical role in embryogenesis and tissue homeostasis, and its deregulation has been associated with tumor growth. The tumor suppressor SuFu inhibits Hh signaling by preventing the nuclear translocation of Gli and suppressing cell proliferation. Regulation of SuFu activity and stability is key to controlling Hh signaling. Here, we unveil SuFu Negating Protein 1 (SNEP1) as a novel Hh target, that enhances the ubiquitination and proteasomal degradation of SuFu and thus promotes Hh signaling. We further show that the E3 ubiquitin ligase LNX1 plays a critical role in the SNEP1-mediated degradation of SuFu. Accordingly, SNEP1 promotes colorectal cancer (CRC) cell proliferation and tumor growth. High levels of SNEP1 are detected in CRC tissues and are well correlated with poor prognosis in CRC patients. Moreover, SNEP1 overexpression reduces sensitivity to anti-Hh inhibitor in CRC cells. Altogether, our findings demonstrate that SNEP1 acts as a novel feedback regulator of Hh signaling by destabilizing SuFu and promoting tumor growth and anti-Hh resistance.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
PGE2-JNK signaling axis non-canonically promotes Gli activation by protecting Gli2 from ubiquitin-proteasomal degradation.Cell Death Dis. 2021 Jul 15;12(7):707. doi: 10.1038/s41419-021-03995-z. Cell Death Dis. 2021. PMID: 34267186 Free PMC article.
-
Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved.Genes Dev. 2009 Aug 15;23(16):1910-28. doi: 10.1101/gad.1794109. Genes Dev. 2009. PMID: 19684112 Free PMC article.
-
Nek2A/SuFu feedback loop regulates Gli-mediated Hedgehog signaling pathway.Int J Oncol. 2017 Feb;50(2):373-380. doi: 10.3892/ijo.2016.3819. Epub 2016 Dec 23. Int J Oncol. 2017. PMID: 28035348 Free PMC article.
-
Regulation of Hh/Gli signaling by dual ubiquitin pathways.Cell Cycle. 2006 Nov 1;5(21):2457-63. doi: 10.4161/cc.5.21.3406. Epub 2006 Sep 14. Cell Cycle. 2006. PMID: 17102630 Review.
-
Role and regulation of human tumor suppressor SUFU in Hedgehog signaling.Adv Cancer Res. 2008;101:29-43. doi: 10.1016/S0065-230X(08)00402-8. Adv Cancer Res. 2008. PMID: 19055941 Review.
Cited by
-
Regulation of Hedgehog Signal Transduction by Ubiquitination and Deubiquitination.Int J Mol Sci. 2021 Dec 11;22(24):13338. doi: 10.3390/ijms222413338. Int J Mol Sci. 2021. PMID: 34948134 Free PMC article. Review.
-
Hedgehog ligand and receptor cooperatively regulate EGFR stability and activity in non-small cell lung cancer.Cell Oncol (Dordr). 2024 Aug;47(4):1405-1423. doi: 10.1007/s13402-024-00938-6. Epub 2024 Apr 3. Cell Oncol (Dordr). 2024. PMID: 38568419
-
Linear polyubiquitylation of Gli protein regulates its protein stability and facilitates tumor growth in colorectal cancer.Cell Death Discov. 2024 Aug 20;10(1):369. doi: 10.1038/s41420-024-02147-4. Cell Death Discov. 2024. PMID: 39164252 Free PMC article.
-
Leptin accelerates BMSC transformation into vertebral epiphyseal plate chondrocytes by activating SENP1-mediated deSUMOylation of SIRT3.FEBS Open Bio. 2023 Feb;13(2):293-306. doi: 10.1002/2211-5463.13539. Epub 2023 Jan 13. FEBS Open Bio. 2023. PMID: 36537765 Free PMC article.
-
Hedgehog pathway and cancer: A new area (Review).Oncol Rep. 2024 Sep;52(3):116. doi: 10.3892/or.2024.8775. Epub 2024 Jul 12. Oncol Rep. 2024. PMID: 38994763 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

