Cerebrospinal fluid proteome shows disrupted neuronal development in multiple sclerosis
- PMID: 33602999
- PMCID: PMC7892850
- DOI: 10.1038/s41598-021-82388-w
Cerebrospinal fluid proteome shows disrupted neuronal development in multiple sclerosis
Abstract
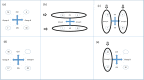
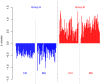
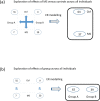
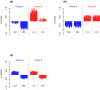
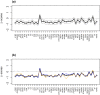
Despite intensive research, the aetiology of multiple sclerosis (MS) remains unknown. Cerebrospinal fluid proteomics has the potential to reveal mechanisms of MS pathogenesis, but analyses must account for disease heterogeneity. We previously reported explorative multivariate analysis by hierarchical clustering of proteomics data of MS patients and controls, which resulted in two groups of individuals. Grouping reflected increased levels of intrathecal inflammatory response proteins and decreased levels of proteins involved in neural development in one group relative to the other group. MS patients and controls were present in both groups. Here we reanalysed these data and we also reanalysed data from an independent cohort of patients diagnosed with clinically isolated syndrome (CIS), who have symptoms of MS without evidence of dissemination in space and/or time. Some, but not all, CIS patients had intrathecal inflammation. The analyses reported here identified a common protein signature of MS/CIS that was not linked to elevated intrathecal inflammation. The signature included low levels of complement proteins, semaphorin-7A, reelin, neural cell adhesion molecules, inter-alpha-trypsin inhibitor heavy chain H2, transforming growth factor beta 1, follistatin-related protein 1, malate dehydrogenase 1 cytoplasmic, plasma retinol-binding protein, biotinidase, and transferrin, all known to play roles in neural development. Low levels of these proteins suggest that MS/CIS patients suffer from abnormally low oxidative capacity that results in disrupted neural development from an early stage of the disease.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
CSF proteome analysis in clinically isolated syndrome (CIS): candidate markers for conversion to definite multiple sclerosis.Neurosci Lett. 2009 Mar 13;452(2):214-7. doi: 10.1016/j.neulet.2009.01.057. Epub 2009 Jan 29. Neurosci Lett. 2009. PMID: 19383442
-
The human CSF pain proteome.J Proteomics. 2019 Jan 6;190:67-76. doi: 10.1016/j.jprot.2018.05.012. Epub 2018 May 28. J Proteomics. 2019. PMID: 29852297
-
Validation of semaphorin 7A and ala-β-his-dipeptidase as biomarkers associated with the conversion from clinically isolated syndrome to multiple sclerosis.J Neuroinflammation. 2014 Nov 13;11:181. doi: 10.1186/s12974-014-0181-8. J Neuroinflammation. 2014. PMID: 25406498 Free PMC article.
-
Insights into the human brain proteome: Disclosing the biological meaning of protein networks in cerebrospinal fluid.Crit Rev Clin Lab Sci. 2017 May;54(3):185-204. doi: 10.1080/10408363.2017.1299682. Epub 2017 Apr 10. Crit Rev Clin Lab Sci. 2017. PMID: 28393582 Review.
-
Oligoclonal bands in multiple sclerosis cerebrospinal fluid: an update on methodology and clinical usefulness.J Neuroimmunol. 2006 Nov;180(1-2):17-28. doi: 10.1016/j.jneuroim.2006.07.006. Epub 2006 Sep 1. J Neuroimmunol. 2006. PMID: 16945427 Review.
Cited by
-
Proteomics in Multiple Sclerosis: The Perspective of the Clinician.Int J Mol Sci. 2022 May 5;23(9):5162. doi: 10.3390/ijms23095162. Int J Mol Sci. 2022. PMID: 35563559 Free PMC article. Review.
-
Quantitative proteomics and multi-omics analysis identifies potential biomarkers and the underlying pathological molecular networks in Chinese patients with multiple sclerosis.BMC Neurol. 2024 Oct 31;24(1):423. doi: 10.1186/s12883-024-03926-3. BMC Neurol. 2024. PMID: 39478468 Free PMC article.
-
Multiple sclerosis and drug discovery: A work of translation.EBioMedicine. 2021 Jun;68:103392. doi: 10.1016/j.ebiom.2021.103392. Epub 2021 May 24. EBioMedicine. 2021. PMID: 34044219 Free PMC article. Review.
-
Identification of brain-enriched proteins in CSF as biomarkers of relapsing remitting multiple sclerosis.Clin Proteomics. 2024 Jun 16;21(1):42. doi: 10.1186/s12014-024-09494-5. Clin Proteomics. 2024. PMID: 38880880 Free PMC article.
-
Proteomics of Multiple Sclerosis: Inherent Issues in Defining the Pathoetiology and Identifying (Early) Biomarkers.Int J Mol Sci. 2021 Jul 9;22(14):7377. doi: 10.3390/ijms22147377. Int J Mol Sci. 2021. PMID: 34298997 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

