The Fate of Speckled Protein 100 (Sp100) During Herpesviruses Infection
- PMID: 33598438
- PMCID: PMC7882683
- DOI: 10.3389/fcimb.2020.607526
The Fate of Speckled Protein 100 (Sp100) During Herpesviruses Infection
Abstract
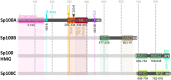
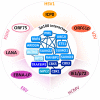
The constitutive expression of Speckled-100 (Sp100) is known to restrict the replication of many clinically important DNA viruses. This pre-existing (intrinsic) immune defense to virus infection can be further upregulated upon interferon (IFN) stimulation as a component of the innate immune response. In humans, Sp100 is encoded by a single gene locus, which can produce alternatively spliced isoforms. The widely studied Sp100A, Sp100B, Sp100C and Sp100HMG have functions associated with the transcriptional regulation of viral and cellular chromatin, either directly through their characteristic DNA-binding domains, or indirectly through post-translational modification (PTM) and associated protein interaction networks. Sp100 isoforms are resident component proteins of promyelocytic leukemia-nuclear bodies (PML-NBs), dynamic nuclear sub-structures which regulate host immune defenses against many pathogens. In the case of human herpesviruses, multiple protein antagonists are expressed to relieve viral DNA genome transcriptional silencing imposed by PML-NB and Sp100-derived proteinaceous structures, thereby stimulating viral propagation, pathogenesis, and transmission to new hosts. This review details how different Sp100 isoforms are manipulated during herpesviruses HSV1, VZV, HCMV, EBV, and KSHV infection, identifying gaps in our current knowledge, and highlighting future areas of research.
Keywords: ISG; PML-NB; Sp100; epigenetics; herpesviruses; immunity.
Copyright © 2021 Collados Rodríguez.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
The interactions between PML nuclear bodies and small and medium size DNA viruses.Virol J. 2023 May 1;20(1):82. doi: 10.1186/s12985-023-02049-4. Virol J. 2023. PMID: 37127643 Free PMC article. Review.
-
Sp100 isoform-specific regulation of human adenovirus 5 gene expression.J Virol. 2014 Jun;88(11):6076-92. doi: 10.1128/JVI.00469-14. Epub 2014 Mar 12. J Virol. 2014. PMID: 24623443 Free PMC article.
-
PML Body Component Sp100A Restricts Wild-Type Herpes Simplex Virus 1 Infection.J Virol. 2022 Apr 27;96(8):e0027922. doi: 10.1128/jvi.00279-22. Epub 2022 Mar 30. J Virol. 2022. PMID: 35353002 Free PMC article.
-
Sp100A promotes chromatin decondensation at a cytomegalovirus-promoter-regulated transcription site.Mol Biol Cell. 2013 May;24(9):1454-68. doi: 10.1091/mbc.E12-09-0669. Epub 2013 Mar 13. Mol Biol Cell. 2013. PMID: 23485562 Free PMC article.
-
A Tale of Usurpation and Subversion: SUMO-Dependent Integrity of Promyelocytic Leukemia Nuclear Bodies at the Crossroad of Infection and Immunity.Front Cell Dev Biol. 2021 Aug 27;9:696234. doi: 10.3389/fcell.2021.696234. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34513832 Free PMC article. Review.
Cited by
-
The extensive transgenerational transcriptomic effects of ocean acidification on the olfactory epithelium of a marine fish are associated with a better viral resistance.BMC Genomics. 2022 Jun 17;23(1):448. doi: 10.1186/s12864-022-08647-w. BMC Genomics. 2022. PMID: 35710351 Free PMC article.
-
The interactions between PML nuclear bodies and small and medium size DNA viruses.Virol J. 2023 May 1;20(1):82. doi: 10.1186/s12985-023-02049-4. Virol J. 2023. PMID: 37127643 Free PMC article. Review.
-
Changes in SUMO-modified proteins in Epstein-Barr virus infection identifies reciprocal regulation of TRIM24/28/33 complexes and the lytic switch BZLF1.PLoS Pathog. 2023 Jul 6;19(7):e1011477. doi: 10.1371/journal.ppat.1011477. eCollection 2023 Jul. PLoS Pathog. 2023. PMID: 37410772 Free PMC article.
-
Functional Roles of Bromodomain Proteins in Cancer.Cancers (Basel). 2021 Jul 19;13(14):3606. doi: 10.3390/cancers13143606. Cancers (Basel). 2021. PMID: 34298819 Free PMC article. Review.
-
It Runs in the Bromodomain Family: Speckled Proteins (SP) Play a Role in the Antitumor Immune Response in Solid Tumors.Int J Mol Sci. 2022 Dec 29;24(1):549. doi: 10.3390/ijms24010549. Int J Mol Sci. 2022. PMID: 36614001 Free PMC article.
References
-
- Aggarwal A., Miranda-Saksena M., Boadle R. A., Kelly B. J., Diefenbach R. J., Alam W., et al. (2012). Ultrastructural visualization of individual tegument protein dissociation during entry of herpes simplex virus 1 into human and rat dorsal root ganglion neurons. J. Virol. 86, 6123–6137. 10.1128/JVI.07016-11 - DOI - PMC - PubMed
-
- Alandijany T., Roberts A. P. E., Conn K. L., Loney C., McFarlane S., Orr A., et al. (2018). Distinct temporal roles for the promyelocytic leukaemia (PML) protein in the sequential regulation of intracellular host immunity to HSV-1 infection. PLoS Pathog. 14, e1006769. 10.1371/journal.ppat.1006769 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

