Supramolecular organization of rhodopsin in rod photoreceptor cell membranes
- PMID: 33591421
- PMCID: PMC8364927
- DOI: 10.1007/s00424-021-02522-5
Supramolecular organization of rhodopsin in rod photoreceptor cell membranes
Abstract
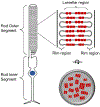
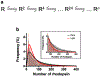
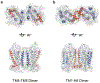
Rhodopsin is the light receptor in rod photoreceptor cells that initiates scotopic vision. Studies on the light receptor span well over a century, yet questions about the organization of rhodopsin within the photoreceptor cell membrane still persist and a consensus view on the topic is still elusive. Rhodopsin has been intensely studied for quite some time, and there is a wealth of information to draw from to formulate an organizational picture of the receptor in native membranes. Early experimental evidence in apparent support for a monomeric arrangement of rhodopsin in rod photoreceptor cell membranes is contrasted and reconciled with more recent visual evidence in support of a supramolecular organization of rhodopsin. What is known so far about the determinants of forming a supramolecular structure and possible functional roles for such an organization are also discussed. Many details are still missing on the structural and functional properties of the supramolecular organization of rhodopsin in rod photoreceptor cell membranes. The emerging picture presented here can serve as a springboard towards a more in-depth understanding of the topic.
Keywords: G protein-coupled receptor; Membrane protein; Nanodomain; Photoreceptor cell; Phototransduction; Quaternary structure.
© 2021. The Author(s), under exclusive licence to Springer-Verlag GmbH, DE part of Springer Nature.
Conflict of interest statement
Conflict of Interest
The author declares that they have no conflicts of interest.
Figures


Similar articles
-
Conservation of molecular interactions stabilizing bovine and mouse rhodopsin.Biochemistry. 2010 Dec 14;49(49):10412-20. doi: 10.1021/bi101345x. Epub 2010 Nov 11. Biochemistry. 2010. PMID: 21038881 Free PMC article.
-
Investigating the Nanodomain Organization of Rhodopsin in Native Membranes by Atomic Force Microscopy.Methods Mol Biol. 2019;1886:61-74. doi: 10.1007/978-1-4939-8894-5_4. Methods Mol Biol. 2019. PMID: 30374862 Free PMC article.
-
Adaptations in rod outer segment disc membranes in response to environmental lighting conditions.Biochim Biophys Acta Mol Cell Res. 2017 Oct;1864(10):1691-1702. doi: 10.1016/j.bbamcr.2017.06.013. Epub 2017 Jun 20. Biochim Biophys Acta Mol Cell Res. 2017. PMID: 28645515 Free PMC article.
-
Rhodopsin Oligomerization and Aggregation.J Membr Biol. 2019 Oct;252(4-5):413-423. doi: 10.1007/s00232-019-00078-1. Epub 2019 Jul 8. J Membr Biol. 2019. PMID: 31286171 Free PMC article. Review.
-
A physiological role for the supramolecular organization of rhodopsin and transducin in rod photoreceptors.FEBS Lett. 2013 Jun 27;587(13):2060-6. doi: 10.1016/j.febslet.2013.05.017. Epub 2013 May 16. FEBS Lett. 2013. PMID: 23684654 Review.
Cited by
-
Where vision begins.Pflugers Arch. 2021 Sep;473(9):1333-1337. doi: 10.1007/s00424-021-02605-3. Pflugers Arch. 2021. PMID: 34245377 Free PMC article. No abstract available.
-
The hypothalamic transcriptome reveals the importance of visual perception on the egg production of Wanxi white geese.Front Vet Sci. 2024 Sep 20;11:1449032. doi: 10.3389/fvets.2024.1449032. eCollection 2024. Front Vet Sci. 2024. PMID: 39372898 Free PMC article.
-
Aggregation of rhodopsin mutants in mouse models of autosomal dominant retinitis pigmentosa.Nat Commun. 2024 Feb 16;15(1):1451. doi: 10.1038/s41467-024-45748-4. Nat Commun. 2024. PMID: 38365903 Free PMC article.
-
Understanding the Rhodopsin Worldview Through Atomic Force Microscopy (AFM): Structure, Stability, and Activity Studies.Chem Rec. 2023 Oct;23(10):e202300113. doi: 10.1002/tcr.202300113. Epub 2023 Jun 2. Chem Rec. 2023. PMID: 37265335 Free PMC article. Review.
References
-
- Albert AD, Young JE, Paw Z (1998) Phospholipid fatty acyl spatial distribution in bovine rod outer segment disk membranes. Biochim Biophys Acta 1368:52–60. doi:S0005-2736(97)00200-9 [pii] - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

