Characterizing the Tumor Immune Microenvironment with Tyramide-Based Multiplex Immunofluorescence
- PMID: 33590360
- PMCID: PMC7960613
- DOI: 10.1007/s10911-021-09479-2
Characterizing the Tumor Immune Microenvironment with Tyramide-Based Multiplex Immunofluorescence
Abstract
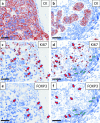
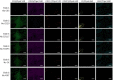
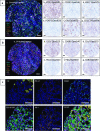
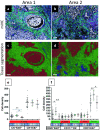
Multiplex immunofluorescence (mIF) allows simultaneous antibody-based detection of multiple markers with a nuclear counterstain on a single tissue section. Recent studies have demonstrated that mIF is becoming an important tool for immune profiling the tumor microenvironment, further advancing our understanding of the interplay between cancer and the immune system, and identifying predictive biomarkers of response to immunotherapy. Expediting mIF discoveries is leading to improved diagnostic panels, whereas it is important that mIF protocols be standardized to facilitate their transition into clinical use. Manual processing of sections for mIF is time consuming and a potential source of variability across numerous samples. To increase reproducibility and throughput we demonstrate the use of an automated slide stainer for mIF incorporating tyramide signal amplification (TSA). We describe two panels aimed at characterizing the tumor immune microenvironment. Panel 1 included CD3, CD20, CD117, FOXP3, Ki67, pancytokeratins (CK), and DAPI, and Panel 2 included CD3, CD8, CD68, PD-1, PD-L1, CK, and DAPI. Primary antibodies were first tested by standard immunohistochemistry and single-plex IF, then multiplex panels were developed and images were obtained using a Vectra 3.0 multispectral imaging system. Various methods for image analysis (identifying cell types, determining cell densities, characterizing cell-cell associations) are outlined. These mIF protocols will be invaluable tools for immune profiling the tumor microenvironment.
Keywords: Breast cancer; Immune cells; Immunohistochemistry; Multiplex.
Figures




Similar articles
-
Multi-institutional TSA-amplified Multiplexed Immunofluorescence Reproducibility Evaluation (MITRE) Study.J Immunother Cancer. 2021 Jul;9(7):e002197. doi: 10.1136/jitc-2020-002197. J Immunother Cancer. 2021. PMID: 34266881 Free PMC article.
-
Validation of multiplex immunofluorescence panels using multispectral microscopy for immune-profiling of formalin-fixed and paraffin-embedded human tumor tissues.Sci Rep. 2017 Oct 17;7(1):13380. doi: 10.1038/s41598-017-13942-8. Sci Rep. 2017. PMID: 29042640 Free PMC article.
-
Immuno-profiling and cellular spatial analysis using five immune oncology multiplex immunofluorescence panels for paraffin tumor tissue.Sci Rep. 2021 Apr 19;11(1):8511. doi: 10.1038/s41598-021-88156-0. Sci Rep. 2021. PMID: 33875760 Free PMC article.
-
Multiplex Immunofluorescence Tyramide Signal Amplification for Immune Cell Profiling of Paraffin-Embedded Tumor Tissues.Front Mol Biosci. 2021 Apr 29;8:667067. doi: 10.3389/fmolb.2021.667067. eCollection 2021. Front Mol Biosci. 2021. PMID: 33996912 Free PMC article. Review.
-
Delineating the breast cancer immune microenvironment in the era of multiplex immunohistochemistry/immunofluorescence.Histopathology. 2021 Aug;79(2):139-159. doi: 10.1111/his.14328. Epub 2021 May 6. Histopathology. 2021. PMID: 33400265 Review.
Cited by
-
BCG as an Innovative Option for HCC Treatment: Repurposing and Mechanistic Insights.Adv Sci (Weinh). 2024 Apr;11(14):e2308242. doi: 10.1002/advs.202308242. Epub 2024 Feb 2. Adv Sci (Weinh). 2024. PMID: 38308164 Free PMC article.
-
Melanoma progression and prognostic models drawn from single-cell, spatial maps of benign and malignant tumors.Sci Adv. 2024 Jul 12;10(28):eadm8206. doi: 10.1126/sciadv.adm8206. Epub 2024 Jul 12. Sci Adv. 2024. PMID: 38996022 Free PMC article.
-
Statistical Analysis of Multiplex Immunofluorescence and Immunohistochemistry Imaging Data.Methods Mol Biol. 2023;2629:141-168. doi: 10.1007/978-1-0716-2986-4_8. Methods Mol Biol. 2023. PMID: 36929077 Review.
-
Atezolizumab plus stereotactic ablative radiotherapy for medically inoperable patients with early-stage non-small cell lung cancer: a multi-institutional phase I trial.Nat Commun. 2023 Sep 2;14(1):5332. doi: 10.1038/s41467-023-40813-w. Nat Commun. 2023. PMID: 37658083 Free PMC article. Clinical Trial.
-
Activated tissue resident memory T-cells (CD8+CD103+CD39+) uniquely predict survival in left sided "immune-hot" colorectal cancers.Front Immunol. 2023 May 11;14:1057292. doi: 10.3389/fimmu.2023.1057292. eCollection 2023. Front Immunol. 2023. PMID: 37251410 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

