Metabolomics Exploration of Pseudorabies Virus Reprogramming Metabolic Profiles of PK-15 Cells to Enhance Viral Replication
- PMID: 33585273
- PMCID: PMC7879706
- DOI: 10.3389/fcimb.2020.599087
Metabolomics Exploration of Pseudorabies Virus Reprogramming Metabolic Profiles of PK-15 Cells to Enhance Viral Replication
Abstract
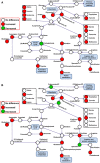
For viral replication to occur in host cells, low-molecular-weight metabolites are necessary for virion assembly. Recently, metabolomics has shown great promise in uncovering the highly complex mechanisms associated with virus-host interactions. In this study, the metabolic networks in PK-15 cells infected with a variant virulent or classical attenuated pseudorabies virus (PRV) strains were explored using gas chromatography-mass spectrometry (GC-MS) analysis. Although total numbers of metabolites whose levels were altered by infection with the variant virulent strain or the classical attenuated strain were different at 8 and 16 h post infection (hpi), the predicted levels of differential metabolic components were shown to be associated with specific pathways, including glycolysis as well as amino acid and nucleotide metabolism. The glucose depletion and glycolysis inhibitors 2DG and oxamate could reduce the level of PRV replication in PK-15 cells. In addition, the inhibition of the pentose phosphate pathway (PPP) resulted in an obvious decline of viral titers, but the prevention of oxidative phosphorylation in the tricarboxylic acid (TCA) cycle had a minimal effect on viral replication. Glutamine starvation resulted in the decline of viral titers, which could be restored by supplemental addition in the culture media. However, inhibition of glutaminase (GLS) activity or the supplement of 2-ketoglutarate into glutamine-deleted DMEM did not alter PRV replication in PK-15 cells. The results of the current study indicate that PRV reprograms the metabolic activities of PK-15 cells. The metabolic flux from glycolysis, PPP and glutamine metabolism to nucleotide biosynthesis was essential for PRV to enhance its replication. This study will help to identify the biochemical materials utilized by PRV replication in host cells, and this knowledge can aid in developing new antiviral strategies.
Keywords: PK-15 cells; classical attenuated strain; metabolic activity; metabolomics; pseudorabies virus (PRV); variant virulent strain.
Copyright © 2021 Gou, Bian, Li, Cai, Jiang, Song, Zhang, Chu, Yang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Metabolomics Analysis of PK-15 Cells with Pseudorabies Virus Infection Based on UHPLC-QE-MS.Viruses. 2022 May 27;14(6):1158. doi: 10.3390/v14061158. Viruses. 2022. PMID: 35746630 Free PMC article.
-
Untargeted LC-MS based metabolomic profiling of iPAMs to investigate lipid metabolic pathways alternations induced by different Pseudorabies virus strains.Vet Microbiol. 2021 May;256:109041. doi: 10.1016/j.vetmic.2021.109041. Epub 2021 Mar 19. Vet Microbiol. 2021. PMID: 33813308
-
Porcine β-defensin 2 inhibits proliferation of pseudorabies virus in vitro and in transgenic mice.Virol J. 2020 Feb 3;17(1):18. doi: 10.1186/s12985-020-1288-4. Virol J. 2020. PMID: 32014007 Free PMC article.
-
Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine.Microbiol Mol Biol Rev. 2005 Sep;69(3):462-500. doi: 10.1128/MMBR.69.3.462-500.2005. Microbiol Mol Biol Rev. 2005. PMID: 16148307 Free PMC article. Review.
-
Pseudorabies (Aujeszky's disease) virus: state of the art. August 1993.Acta Vet Hung. 1994;42(2-3):153-77. Acta Vet Hung. 1994. PMID: 7810409 Review.
Cited by
-
Lactate is useful for the efficient replication of porcine epidemic diarrhea virus in cell culture.Front Vet Sci. 2023 Feb 13;10:1116695. doi: 10.3389/fvets.2023.1116695. eCollection 2023. Front Vet Sci. 2023. PMID: 36861007 Free PMC article.
-
Metabolomics Analysis of PK-15 Cells with Pseudorabies Virus Infection Based on UHPLC-QE-MS.Viruses. 2022 May 27;14(6):1158. doi: 10.3390/v14061158. Viruses. 2022. PMID: 35746630 Free PMC article.
-
Genomic regions associated with pseudorabies virus infection status in naturally infected feral swine (Sus scrofa).Front Genet. 2023 Nov 23;14:1292671. doi: 10.3389/fgene.2023.1292671. eCollection 2023. Front Genet. 2023. PMID: 38075681 Free PMC article.
-
RIPK3-Dependent Necroptosis Limits PRV Replication in PK-15 Cells.Front Microbiol. 2021 Jun 4;12:664353. doi: 10.3389/fmicb.2021.664353. eCollection 2021. Front Microbiol. 2021. PMID: 34149651 Free PMC article.
-
Cellular metabolism hijacked by viruses for immunoevasion: potential antiviral targets.Front Immunol. 2023 Jul 25;14:1228811. doi: 10.3389/fimmu.2023.1228811. eCollection 2023. Front Immunol. 2023. PMID: 37559723 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

