Next-Generation Sequencing and the CRISPR-Cas Nexus: A Molecular Plant Virology Perspective
- PMID: 33584572
- PMCID: PMC7874184
- DOI: 10.3389/fmicb.2020.609376
Next-Generation Sequencing and the CRISPR-Cas Nexus: A Molecular Plant Virology Perspective
Abstract
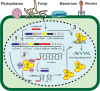
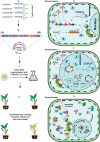
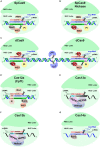
In recent years, next-generation sequencing (NGS) and contemporary Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-CRISPR-associated (Cas) technologies have revolutionized the life sciences and the field of plant virology. Both these technologies offer an unparalleled platform for sequencing and deciphering viral metagenomes promptly. Over the past two decades, NGS technologies have improved enormously and have impacted plant virology. NGS has enabled the detection of plant viruses that were previously undetectable by conventional approaches, such as quarantine and archeological plant samples, and has helped to track the evolutionary footprints of viral pathogens. The CRISPR-Cas-based genome editing (GE) and detection techniques have enabled the development of effective approaches to virus resistance. Different versions of CRISPR-Cas have been employed to successfully confer resistance against diverse plant viruses by directly targeting the virus genome or indirectly editing certain host susceptibility factors. Applications of CRISPR-Cas systems include targeted insertion and/or deletion, site-directed mutagenesis, induction/expression/repression of the gene(s), epigenome re-modeling, and SNPs detection. The CRISPR-Cas toolbox has been equipped with precision GE tools to engineer the target genome with and without double-stranded (ds) breaks or donor templates. This technique has also enabled the generation of transgene-free genetically engineered plants, DNA repair, base substitution, prime editing, detection of small molecules, and biosensing in plant virology. This review discusses the utilities, advantages, applications, bottlenecks of NGS, and CRISPR-Cas in plant virology.
Keywords: CRISPR; CRISPR associated (Cas) proteins; genome editing; next generation sequencing (NGS); plant viruses.
Copyright © 2021 Shahid, Sattar, Iqbal, Raza and Al-Sadi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Integrating CRISPR-Cas and Next Generation Sequencing in Plant Virology.Front Genet. 2021 Oct 25;12:735489. doi: 10.3389/fgene.2021.735489. eCollection 2021. Front Genet. 2021. PMID: 34759957 Free PMC article. Review.
-
CRISPER/Cas in Plant Natural Product Research: Therapeutics as Anticancer and other Drug Candidates and Recent Patents.Recent Pat Anticancer Drug Discov. 2021;16(4):460-468. doi: 10.2174/1574892816666210706155602. Recent Pat Anticancer Drug Discov. 2021. PMID: 34911411 Review.
-
Next-Generation Sequencing and Genome Editing in Plant Virology.Front Microbiol. 2016 Aug 26;7:1325. doi: 10.3389/fmicb.2016.01325. eCollection 2016. Front Microbiol. 2016. PMID: 27617007 Free PMC article. Review.
-
Tools and targets: The dual role of plant viruses in CRISPR-Cas genome editing.Plant Genome. 2023 Jun;16(2):e20220. doi: 10.1002/tpg2.20220. Epub 2022 Jun 14. Plant Genome. 2023. PMID: 35698891 Review.
-
CRISPR Applications in Plant Virology: Virus Resistance and Beyond.Phytopathology. 2020 Jan;110(1):18-28. doi: 10.1094/PHYTO-07-19-0267-IA. Epub 2019 Nov 13. Phytopathology. 2020. PMID: 31433273 Review.
Cited by
-
Clustered Regularly Interspaced Short Palindromic Repeats-Associated Protein System for Resistance Against Plant Viruses: Applications and Perspectives.Front Plant Sci. 2022 May 26;13:904829. doi: 10.3389/fpls.2022.904829. eCollection 2022. Front Plant Sci. 2022. PMID: 35693174 Free PMC article. Review.
-
High-throughput sequencing discovered diverse monopartite and bipartite begomoviruses infecting cucumbers in Saudi Arabia.Front Plant Sci. 2024 Oct 10;15:1375405. doi: 10.3389/fpls.2024.1375405. eCollection 2024. Front Plant Sci. 2024. PMID: 39450090 Free PMC article.
-
Coinfection of Two Mycoviruses Confers Hypovirulence and Reduces the Production of Mycotoxin Alternariol in Alternaria alternata f. sp. mali.Front Microbiol. 2022 Jun 9;13:910712. doi: 10.3389/fmicb.2022.910712. eCollection 2022. Front Microbiol. 2022. PMID: 35756001 Free PMC article.
-
High-Throughput Sequencing Identified Distinct Bipartite and Monopartite Begomovirus Variants Associated with DNA-Satellites from Tomato and Muskmelon Plants in Saudi Arabia.Plants (Basel). 2022 Dec 20;12(1):6. doi: 10.3390/plants12010006. Plants (Basel). 2022. PMID: 36616136 Free PMC article.
-
A Loop-Mediated Isothermal Amplification Assay for the Rapid Detection of Didymella segeticola Causing Tea Leaf Spot.J Fungi (Basel). 2024 Jul 3;10(7):467. doi: 10.3390/jof10070467. J Fungi (Basel). 2024. PMID: 39057352 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

