The role of ARHGAP9: clinical implication and potential function in acute myeloid leukemia
- PMID: 33579308
- PMCID: PMC7881617
- DOI: 10.1186/s12967-021-02733-5
The role of ARHGAP9: clinical implication and potential function in acute myeloid leukemia
Abstract
Background: Rho GTPase activating protein 9 (ARHGAP9) is expressed in various types of cancers and can inactivate Rho GTPases that mainly regulate cytoskeletal dynamics. However, the exact role of ARHGAP9 in acute myeloid leukemia (AML) has yet to be clarified.
Methods: We compared the transcriptional expression, prognosis, differentially expressed genes, functional enrichment, and hub genes in AML patients on the basis of the data published in the following databases: UALCAN, GEPIA, Gene Expression Omnibus, the Human Protein Atlas, Cancer Cell Line Encyclopedia, LinkedOmics, Metascape, and String. Data from the Cancer Genome Atlas database was used to evaluate the correlations between ARHGAP9 expression and various clinicopathological parameters, as well as the significantly different genes associated with ARHGAP9 expression.
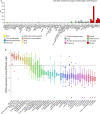
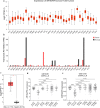
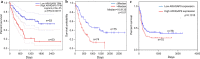
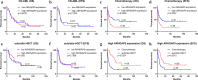
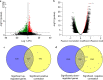
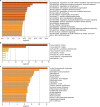
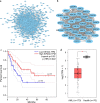
Results: We found that ARHGAP9 expression was higher in the tissues and cell lines extracted from patients with AML than corresponding control tissues and other cancer types. ARHGAP9 overexpression was associated with decreased overall survival (OS) in AML. Compared with the ARHGAP9low group, the ARHGAP9high group, which received only chemotherapy, showed significantly worse OS and event-free survival (EFS); however, no significant difference was observed after treatment with autologous or allogeneic hematopoietic stem cell transplantation (auto/allo-HSCT). The ARHGAP9high patients undergoing auto/allo-HSCT also had a significantly better prognosis with respect to OS and EFS than those receiving only chemotherapy. Most overlapping genes of the significantly different genes and co-expression genes exhibited enriched immune functions, suggesting the immune regulation potential of ARHGAP9 in AML. A total of 32 hub genes were identified from the differentially expressed genes, within which the KIF20A had a significant prognostic value for AML.
Conclusions: ARHGAP9 overexpression was associated with poor OS in AML patients and can be used as a prognostic biomarker. AML patients with ARHGAP9 overexpression can benefit from auto/allo-HSCT rather than chemotherapy.
Keywords: AML; ARHGAP9; Auto/allo-HSCT; Chemotherapy; Prognosis; t(15;17).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
BCL2 overexpression: clinical implication and biological insights in acute myeloid leukemia.Diagn Pathol. 2019 Jun 29;14(1):68. doi: 10.1186/s13000-019-0841-1. Diagn Pathol. 2019. PMID: 31253168 Free PMC article.
-
The prognostic value of RASGEF1A RNA expression and DNA methylation in cytogenetically normal acute myeloid leukemia.Cancer Biomark. 2023;36(2):103-116. doi: 10.3233/CBM-210407. Cancer Biomark. 2023. PMID: 36404533
-
High expression of AK1 predicts inferior prognosis in acute myeloid leukemia patients undergoing chemotherapy.Biosci Rep. 2020 Jun 26;40(6):BSR20200097. doi: 10.1042/BSR20200097. Biosci Rep. 2020. PMID: 32519744 Free PMC article.
-
Is allogeneic transplantation really the best treatment for FLT3/ITD-positive acute myeloid leukemia? A systematic review.Clin Transplant. 2015 Feb;29(2):149-60. doi: 10.1111/ctr.12495. Epub 2015 Jan 2. Clin Transplant. 2015. PMID: 25430616 Review.
-
Rho GTPase Activating Protein 9 (ARHGAP9) in Human Cancers.Recent Pat Anticancer Drug Discov. 2022;17(1):55-65. doi: 10.2174/1574892816666210806155754. Recent Pat Anticancer Drug Discov. 2022. PMID: 34365932 Review.
Cited by
-
Clinical implication and potential function of ARHGEF6 in acute myeloid leukemia: An in vitro study.PLoS One. 2023 Apr 7;18(4):e0283934. doi: 10.1371/journal.pone.0283934. eCollection 2023. PLoS One. 2023. PMID: 37027440 Free PMC article.
-
CDK19 regulates the proliferation of hematopoietic stem cells and acute myeloid leukemia cells by suppressing p53-mediated transcription of p21.Leukemia. 2022 Apr;36(4):956-969. doi: 10.1038/s41375-022-01512-5. Epub 2022 Feb 2. Leukemia. 2022. PMID: 35110726
-
Fixing the GAP: The role of RhoGAPs in cancer.Eur J Cell Biol. 2022 Apr;101(2):151209. doi: 10.1016/j.ejcb.2022.151209. Epub 2022 Feb 10. Eur J Cell Biol. 2022. PMID: 35180567 Free PMC article. Review.
-
Identification of NAD+ Metabolism-Derived Gene Signatures in Ovarian Cancer Prognosis and Immunotherapy.Front Genet. 2022 Jun 16;13:905238. doi: 10.3389/fgene.2022.905238. eCollection 2022. Front Genet. 2022. PMID: 35783253 Free PMC article.
-
GATA binding protein 5 (GATA5) induces Rho GTPase activating protein 9 (ARHGAP9) to inhibit the malignant process of lung adenocarcinoma cells.Bioengineered. 2022 Feb;13(2):2878-2888. doi: 10.1080/21655979.2022.2025695. Bioengineered. 2022. PMID: 35040754 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

