Hepatitis B virus integrations promote local and distant oncogenic driver alterations in hepatocellular carcinoma
- PMID: 33563643
- PMCID: PMC8862055
- DOI: 10.1136/gutjnl-2020-323153
Hepatitis B virus integrations promote local and distant oncogenic driver alterations in hepatocellular carcinoma
Abstract
Objective: Infection by HBV is the main risk factor for hepatocellular carcinoma (HCC) worldwide. HBV directly drives carcinogenesis through integrations in the human genome. This study aimed to precisely characterise HBV integrations, in relation with viral and host genomics and clinical features.
Design: A novel pipeline was set up to perform viral capture on tumours and non-tumour liver tissues from a French cohort of 177 patients mainly of European and African origins. Clonality of each integration event was determined with the localisation, orientation and content of the integrated sequence. In three selected tumours, complex integrations were reconstructed using long-read sequencing or Bionano whole genome mapping.
Results: Replicating HBV DNA was more frequently detected in non-tumour tissues and associated with a higher number of non-clonal integrations. In HCC, clonal selection of HBV integrations was related to two different mechanisms involved in carcinogenesis. First, integration of viral enhancer nearby a cancer-driver gene may lead to a strong overexpression of oncogenes. Second, we identified frequent chromosome rearrangements at HBV integration sites leading to cancer-driver genes (TERT, TP53, MYC) alterations at distance. Moreover, HBV integrations have direct clinical implications as HCC with a high number of insertions develop in young patients and have a poor prognosis.
Conclusion: Deep characterisation of HBV integrations in liver tissues highlights new HBV-associated driver mechanisms involved in hepatocarcinogenesis. HBV integrations have multiple direct oncogenic consequences that remain an important challenge for the follow-up of HBV-infected patients.
Keywords: cancer genetics; hepatitis B; hepatocellular carcinoma.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures



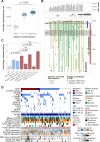
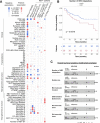
Comment in
-
Genome-wide analysis of hepatitis B virus integration in hepatocellular carcinoma: Insights next generation sequencing.Hepatobiliary Surg Nutr. 2021 Aug;10(4):548-552. doi: 10.21037/hbsn-21-228. Hepatobiliary Surg Nutr. 2021. PMID: 34430541 Free PMC article. No abstract available.
Similar articles
-
HBV integrations reshaping genomic structures promote hepatocellular carcinoma.Gut. 2024 Jun 6;73(7):1169-1182. doi: 10.1136/gutjnl-2023-330414. Gut. 2024. PMID: 38395437 Free PMC article.
-
Targeted Long-Read Sequencing Reveals Comprehensive Architecture, Burden, and Transcriptional Signatures from Hepatitis B Virus-Associated Integrations and Translocations in Hepatocellular Carcinoma Cell Lines.J Virol. 2021 Sep 9;95(19):e0029921. doi: 10.1128/JVI.00299-21. Epub 2021 Jul 21. J Virol. 2021. PMID: 34287049 Free PMC article.
-
Characterization of Hepatitis B Virus Integrations Identified in Hepatocellular Carcinoma Genomes.Viruses. 2021 Feb 4;13(2):245. doi: 10.3390/v13020245. Viruses. 2021. PMID: 33557409 Free PMC article.
-
Hepadnaviruses in cirrhotic liver and hepatocellular carcinoma.J Med Virol. 1990 May;31(1):18-32. doi: 10.1002/jmv.1890310106. J Med Virol. 1990. PMID: 2165515 Review.
-
Hepatitis B Virus DNA Integration Drives Carcinogenesis and Provides a New Biomarker for HBV-related HCC.Cell Mol Gastroenterol Hepatol. 2023;15(4):921-929. doi: 10.1016/j.jcmgh.2023.01.001. Epub 2023 Jan 20. Cell Mol Gastroenterol Hepatol. 2023. PMID: 36690297 Free PMC article. Review.
Cited by
-
Analysis of viral integration reveals new insights of oncogenic mechanism in HBV-infected intrahepatic cholangiocarcinoma and combined hepatocellular-cholangiocarcinoma.Hepatol Int. 2022 Dec;16(6):1339-1352. doi: 10.1007/s12072-022-10419-3. Epub 2022 Sep 20. Hepatol Int. 2022. PMID: 36123506 Free PMC article.
-
Targeted long-read sequencing reveals clonally expanded HBV-associated chromosomal translocations in patients with chronic hepatitis B.JHEP Rep. 2022 Feb 12;4(4):100449. doi: 10.1016/j.jhepr.2022.100449. eCollection 2022 Apr. JHEP Rep. 2022. PMID: 35295767 Free PMC article.
-
Tenofovir alafenamide and tenofovir disoproxil fumarate reduce incidence of hepatocellular carcinoma in patients with chronic hepatitis B.JHEP Rep. 2023 Jul 13;5(10):100847. doi: 10.1016/j.jhepr.2023.100847. eCollection 2023 Oct. JHEP Rep. 2023. PMID: 37771546 Free PMC article.
-
Application of third-generation sequencing in cancer research.Med Rev (2021). 2021 Oct 21;1(2):150-171. doi: 10.1515/mr-2021-0013. eCollection 2021 Dec. Med Rev (2021). 2021. PMID: 37724303 Free PMC article.
-
Serum resistin and the risk for hepatocellular carcinoma in diabetic patients.World J Gastroenterol. 2023 Jul 21;29(27):4271-4288. doi: 10.3748/wjg.v29.i27.4271. World J Gastroenterol. 2023. PMID: 37545641 Free PMC article. Review.
References
-
- World Health Organization . Global hepatitis report, 2017; 2017.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
