Single-molecule force spectroscopy reveals the dynamic strength of the hair-cell tip-link connection
- PMID: 33558532
- PMCID: PMC7870652
- DOI: 10.1038/s41467-021-21033-6
Single-molecule force spectroscopy reveals the dynamic strength of the hair-cell tip-link connection
Abstract
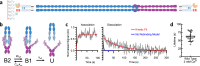
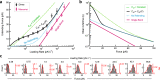
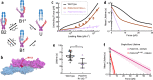
The conversion of auditory and vestibular stimuli into electrical signals is initiated by force transmitted to a mechanotransduction channel through the tip link, a double stranded protein filament held together by two adhesion bonds in the middle. Although thought to form a relatively static structure, the dynamics of the tip-link connection has not been measured. Here, we biophysically characterize the strength of the tip-link connection at single-molecule resolution. We show that a single tip-link bond is more mechanically stable relative to classic cadherins, and our data indicate that the double stranded tip-link connection is stabilized by single strand rebinding facilitated by strong cis-dimerization domains. The measured lifetime of seconds suggests the tip-link is far more dynamic than previously thought. We also show how Ca2+ alters tip-link lifetime through elastic modulation and reveal the mechanical phenotype of a hereditary deafness mutation. Together, these data show how the tip link is likely to function during mechanical stimuli.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells.Nature. 2007 Sep 6;449(7158):87-91. doi: 10.1038/nature06091. Nature. 2007. PMID: 17805295
-
Structure of a force-conveying cadherin bond essential for inner-ear mechanotransduction.Nature. 2012 Dec 6;492(7427):128-32. doi: 10.1038/nature11590. Epub 2012 Nov 7. Nature. 2012. PMID: 23135401 Free PMC article.
-
Cadherins and mechanotransduction by hair cells.Curr Opin Cell Biol. 2008 Oct;20(5):557-66. doi: 10.1016/j.ceb.2008.06.004. Epub 2008 Jul 30. Curr Opin Cell Biol. 2008. PMID: 18619539 Free PMC article. Review.
-
Noddy, a mouse harboring a missense mutation in protocadherin-15, reveals the impact of disrupting a critical interaction site between tip-link cadherins in inner ear hair cells.J Neurosci. 2013 Mar 6;33(10):4395-404. doi: 10.1523/JNEUROSCI.4514-12.2013. J Neurosci. 2013. PMID: 23467356 Free PMC article.
-
Tip links in hair cells: molecular composition and role in hearing loss.Curr Opin Otolaryngol Head Neck Surg. 2009 Oct;17(5):388-93. doi: 10.1097/MOO.0b013e3283303472. Curr Opin Otolaryngol Head Neck Surg. 2009. PMID: 19633555 Free PMC article. Review.
Cited by
-
Anisotropy in mechanical unfolding of protein upon partner-assisted pulling and handle-assisted pulling.Commun Biol. 2021 Jul 29;4(1):925. doi: 10.1038/s42003-021-02445-y. Commun Biol. 2021. PMID: 34326473 Free PMC article.
-
Sensing sound: Cellular specializations and molecular force sensors.Neuron. 2022 Nov 16;110(22):3667-3687. doi: 10.1016/j.neuron.2022.09.018. Epub 2022 Oct 11. Neuron. 2022. PMID: 36223766 Free PMC article. Review.
-
Direct observation of the conformational states of PIEZO1.Nature. 2023 Aug;620(7976):1117-1125. doi: 10.1038/s41586-023-06427-4. Epub 2023 Aug 16. Nature. 2023. PMID: 37587339 Free PMC article.
-
Mini-PCDH15 gene therapy rescues hearing in a mouse model of Usher syndrome type 1F.Nat Commun. 2023 Apr 26;14(1):2400. doi: 10.1038/s41467-023-38038-y. Nat Commun. 2023. PMID: 37100771 Free PMC article.
-
Mechanotransduction in mammalian sensory hair cells.Mol Cell Neurosci. 2022 May;120:103706. doi: 10.1016/j.mcn.2022.103706. Epub 2022 Feb 23. Mol Cell Neurosci. 2022. PMID: 35218890 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

