The thymoproteasome hardwires the TCR repertoire of CD8+ T cells in the cortex independent of negative selection
- PMID: 33555295
- PMCID: PMC7873839
- DOI: 10.1084/jem.20201904
The thymoproteasome hardwires the TCR repertoire of CD8+ T cells in the cortex independent of negative selection
Abstract
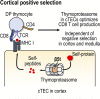
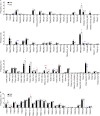
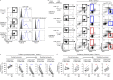
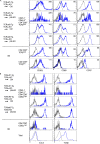
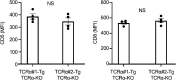
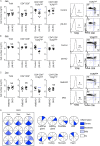
The thymoproteasome expressed specifically in thymic cortical epithelium optimizes the generation of CD8+ T cells; however, how the thymoproteasome contributes to CD8+ T cell development is unclear. Here, we show that the thymoproteasome shapes the TCR repertoire directly in cortical thymocytes before migration to the thymic medulla. We further show that the thymoproteasome optimizes CD8+ T cell production independent of the thymic medulla; independent of additional antigen-presenting cells, including medullary thymic epithelial cells and dendritic cells; and independent of apoptosis-mediated negative selection. These results indicate that the thymoproteasome hardwires the TCR repertoire of CD8+ T cells with cortical positive selection independent of negative selection in the thymus.
© 2021 Ohigashi et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures






Similar articles
-
Thymoproteasome optimizes positive selection of CD8+ T cells without contribution of negative selection.Adv Immunol. 2021;149:1-23. doi: 10.1016/bs.ai.2021.03.001. Epub 2021 May 1. Adv Immunol. 2021. PMID: 33993918 Free PMC article. Review.
-
Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells.Immunity. 2010 Jan 29;32(1):29-40. doi: 10.1016/j.immuni.2009.10.009. Epub 2009 Dec 31. Immunity. 2010. PMID: 20045355
-
β5t-containing thymoproteasome: specific expression in thymic cortical epithelial cells and role in positive selection of CD8+ T cells.Curr Opin Immunol. 2012 Feb;24(1):92-8. doi: 10.1016/j.coi.2012.01.006. Epub 2012 Jan 27. Curr Opin Immunol. 2012. PMID: 22285892 Review.
-
TCR affinity for thymoproteasome-dependent positively selecting peptides conditions antigen responsiveness in CD8(+) T cells.Nat Immunol. 2015 Oct;16(10):1069-76. doi: 10.1038/ni.3237. Epub 2015 Aug 24. Nat Immunol. 2015. PMID: 26301566 Free PMC article.
-
Restricted Expression of the Thymoproteasome Is Required for Thymic Selection and Peripheral Homeostasis of CD8+ T Cells.Cell Rep. 2019 Jan 15;26(3):639-651.e2. doi: 10.1016/j.celrep.2018.12.078. Cell Rep. 2019. PMID: 30650357
Cited by
-
Tissue-specific proteasomes in generation of MHC class I peptides and CD8+ T cells.Curr Opin Immunol. 2022 Aug;77:102217. doi: 10.1016/j.coi.2022.102217. Epub 2022 Jun 8. Curr Opin Immunol. 2022. PMID: 35689940 Free PMC article. Review.
-
Know thy immune self and non-self: Proteomics informs on the expanse of self and non-self, and how and where they arise.Proteomics. 2021 Dec;21(23-24):e2000143. doi: 10.1002/pmic.202000143. Epub 2021 Aug 9. Proteomics. 2021. PMID: 34310018 Free PMC article. Review.
-
The thymoproteasome in shaping the CD8+ T-cell repertoire.Curr Opin Immunol. 2023 Aug;83:102336. doi: 10.1016/j.coi.2023.102336. Epub 2023 May 19. Curr Opin Immunol. 2023. PMID: 37210932 Free PMC article. Review.
-
Peptides for T cell selection in the thymus.Peptides. 2021 Dec;146:170671. doi: 10.1016/j.peptides.2021.170671. Epub 2021 Oct 5. Peptides. 2021. PMID: 34624431 Free PMC article. Review.
-
PTEN directs developmental and metabolic signaling for innate-like T cell fate and tissue homeostasis.Nat Cell Biol. 2022 Nov;24(11):1642-1654. doi: 10.1038/s41556-022-01011-w. Epub 2022 Oct 27. Nat Cell Biol. 2022. PMID: 36302969 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

