Development and External Validation of a Novel Immune Checkpoint-Related Gene Signature for Prediction of Overall Survival in Hepatocellular Carcinoma
- PMID: 33553243
- PMCID: PMC7859359
- DOI: 10.3389/fmolb.2020.620765
Development and External Validation of a Novel Immune Checkpoint-Related Gene Signature for Prediction of Overall Survival in Hepatocellular Carcinoma
Abstract
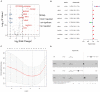
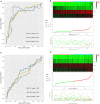
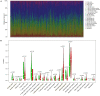
Objective: The purpose of this study was to develop and validate a novel immune checkpoint-related gene signature for prediction of overall survival (OS) in hepatocellular carcinoma (HCC). Methods: mRNA expression profiles and clinical follow-up information were obtained in the International Cancer Genome Consortium database. An external dataset from The Cancer Genome Atlas (TCGA) Liver Hepatocellular Carcinoma database was used to validate the results. The univariate and multivariate Cox regression analyses were performed based on the differentially expressed genes. We generated a four-mRNA signature to predict patient survival. Furthermore, the reliability and validity were validated in TCGA cohort. An integrated bioinformatics approach was performed to evaluate its diagnostic and prognostic value. Results: A four-gene (epidermal growth factor, mutated in colorectal cancer, mitogen-activated protein kinase kinase 2, and NRAS proto-oncogene, GTPase) signature was built to classify patients into two risk groups using a risk score with different OS in two cohorts (all P < 0.0001). Multivariate regression analysis demonstrated the signature was an independent predictor of HCC. Furthermore, the signature presented an excellent diagnostic power in differentiating HCC and adjacent tissues. Immune cell infiltration analysis revealed that the signature was associated with a number of immune cell subtypes. Conclusion: We identified a four-immune checkpoint-related gene signature as a robust biomarker with great potential for clinical application in risk stratification and OS prediction in HCC patients and could be a potential indicator of immunotherapy in HCC. The diagnostic signature had been validated to accurately distinguish HCC from adjacent tissues.
Keywords: database; hepatocellular carcinoma; immune checkpoint; overall survival; signature.
Copyright © 2021 Zhao, Chen and Dang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Identification of a prognostic and therapeutic immune signature associated with hepatocellular carcinoma.Cancer Cell Int. 2021 Feb 10;21(1):98. doi: 10.1186/s12935-021-01792-4. Cancer Cell Int. 2021. PMID: 33568167 Free PMC article.
-
Development and validation of a novel immune-related prognostic model in hepatocellular carcinoma.J Transl Med. 2020 Feb 11;18(1):67. doi: 10.1186/s12967-020-02255-6. J Transl Med. 2020. PMID: 32046766 Free PMC article.
-
Development and validation of a novel pseudogene pair-based prognostic signature for prediction of overall survival in patients with hepatocellular carcinoma.BMC Cancer. 2020 Sep 16;20(1):887. doi: 10.1186/s12885-020-07391-2. BMC Cancer. 2020. PMID: 32938429 Free PMC article.
-
Integrated bioinformatics analyses of key genes involved in hepatocellular carcinoma immunosuppression.Oncol Lett. 2021 Dec;22(6):830. doi: 10.3892/ol.2021.13091. Epub 2021 Oct 13. Oncol Lett. 2021. PMID: 34691257 Free PMC article.
-
Comprehensive analysis reveals a metabolic ten-gene signature in hepatocellular carcinoma.PeerJ. 2020 May 26;8:e9201. doi: 10.7717/peerj.9201. eCollection 2020. PeerJ. 2020. PMID: 32518728 Free PMC article.
Cited by
-
Identification of clinical prognostic factors and analysis of ferroptosis-related gene signatures in the bladder cancer immune microenvironment.BMC Urol. 2024 Jan 3;24(1):6. doi: 10.1186/s12894-023-01354-y. BMC Urol. 2024. PMID: 38172792 Free PMC article.
-
Pyrimidine metabolism regulator-mediated molecular subtypes display tumor microenvironmental hallmarks and assist precision treatment in bladder cancer.Front Oncol. 2023 Aug 17;13:1102518. doi: 10.3389/fonc.2023.1102518. eCollection 2023. Front Oncol. 2023. PMID: 37664033 Free PMC article.
-
Glutamine metabolism genes prognostic signature for stomach adenocarcinoma and immune infiltration: potential biomarkers for predicting overall survival.Front Oncol. 2023 Jun 12;13:1201297. doi: 10.3389/fonc.2023.1201297. eCollection 2023. Front Oncol. 2023. PMID: 37377916 Free PMC article.
-
Novel necroptosis-related gene signature for predicting the prognosis of pancreatic adenocarcinoma.Aging (Albany NY). 2022 Jan 24;14(2):869-891. doi: 10.18632/aging.203846. Epub 2022 Jan 24. Aging (Albany NY). 2022. PMID: 35077391 Free PMC article.
-
Relationship between serum Th1/Th2 imbalance and depression in elderly patients with COPD and its clinical implications.Technol Health Care. 2023;31(6):2047-2058. doi: 10.3233/THC-230665. Technol Health Care. 2023. PMID: 37694327 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

