Dynamics of binding ability prediction between spike protein and human ACE2 reveals the adaptive strategy of SARS-CoV-2 in humans
- PMID: 33542420
- PMCID: PMC7862608
- DOI: 10.1038/s41598-021-82938-2
Dynamics of binding ability prediction between spike protein and human ACE2 reveals the adaptive strategy of SARS-CoV-2 in humans
Abstract
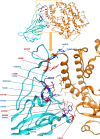
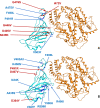
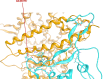
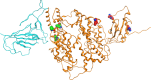
SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) is a novel coronavirus causing the COVID-19 pandemic in 2020. High adaptive plasticity on the spike protein of SASR-CoV-2 enables it to transmit across different host species. In the present study, we collected 2092 high-quality genome sequences of SARS-CoV-2 from 160 regions in over 50 countries and reconstructed their phylogeny. We also analyzed the polymorphic interaction between spike protein and human ACE2 (hACE2). Phylogenetic analysis of SARS-CoV-2 suggests that SARS-CoV-2 is probably originated from a recombination event on the spike protein between a bat coronavirus and a pangolin coronavirus that endows it humans infectivity. Compared with other regions in the S gene of SARS-CoV-2, the direct-binding sites of the receptor-binding domain (RBD) is more conserved. We focused on 3,860 amino acid mutations in spike protein RBD (T333-C525) of SARS-CoV-2 and simulated their differential stability and binding affinity to hACE2 (S19-D615). The results indicate no preference for SARS-CoV-2 infectivity on people of different ethnic groups. The variants in the spike protein of SARS-CoV-2 may also be a good indicator demonstrating the transmission route of SARS-CoV-2 from its natural reservoir to human hosts.
Figures



Similar articles
-
V367F Mutation in SARS-CoV-2 Spike RBD Emerging during the Early Transmission Phase Enhances Viral Infectivity through Increased Human ACE2 Receptor Binding Affinity.J Virol. 2021 Jul 26;95(16):e0061721. doi: 10.1128/JVI.00617-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34105996 Free PMC article.
-
Impact of Genetic Variability in ACE2 Expression on the Evolutionary Dynamics of SARS-CoV-2 Spike D614G Mutation.Genes (Basel). 2020 Dec 24;12(1):16. doi: 10.3390/genes12010016. Genes (Basel). 2020. PMID: 33374416 Free PMC article.
-
Mutations derived from horseshoe bat ACE2 orthologs enhance ACE2-Fc neutralization of SARS-CoV-2.PLoS Pathog. 2021 Apr 9;17(4):e1009501. doi: 10.1371/journal.ppat.1009501. eCollection 2021 Apr. PLoS Pathog. 2021. PMID: 33836016 Free PMC article.
-
Structural basis of severe acute respiratory syndrome coronavirus 2 infection.Curr Opin HIV AIDS. 2021 Jan;16(1):74-81. doi: 10.1097/COH.0000000000000658. Curr Opin HIV AIDS. 2021. PMID: 33186231 Review.
-
Molecular Dynamics Studies on the Structural Characteristics for the Stability Prediction of SARS-CoV-2.Int J Mol Sci. 2021 Aug 13;22(16):8714. doi: 10.3390/ijms22168714. Int J Mol Sci. 2021. PMID: 34445414 Free PMC article. Review.
Cited by
-
Comparison of Clinical Features and Outcomes of Medically Attended COVID-19 and Influenza Patients in a Defined Population in the 2020 Respiratory Virus Season.Front Public Health. 2021 Mar 23;9:587425. doi: 10.3389/fpubh.2021.587425. eCollection 2021. Front Public Health. 2021. PMID: 33834012 Free PMC article.
-
Linking COVID-19 and Heme-Driven Pathophysiologies: A Combined Computational-Experimental Approach.Biomolecules. 2021 Apr 27;11(5):644. doi: 10.3390/biom11050644. Biomolecules. 2021. PMID: 33925394 Free PMC article.
-
A scalable and high yielding SARS-CoV-2 spike protein receptor binding domain production process.Protein Expr Purif. 2023 May;205:106241. doi: 10.1016/j.pep.2023.106241. Epub 2023 Feb 1. Protein Expr Purif. 2023. PMID: 36736512 Free PMC article.
-
Diagnostics of SARS-CoV-2 infection using electrical impedance spectroscopy with an immunosensor to detect the spike protein.Talanta. 2022 Mar 1;239:123076. doi: 10.1016/j.talanta.2021.123076. Epub 2021 Nov 22. Talanta. 2022. PMID: 34876273 Free PMC article.
-
The evolution of SARS-CoV-2 and the COVID-19 pandemic.PeerJ. 2023 Sep 7;11:e15990. doi: 10.7717/peerj.15990. eCollection 2023. PeerJ. 2023. PMID: 37701824 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- 2019BP0202/Project of Basic Research Fund of Henan Institute of Medical and Pharmacological Sciences
- 182102310094/the Key Scientific and Technological Research Projects in Henan Province of China
- 18XTZX12004/the Collaborative Innovation Project of Zhengzhou
- 201100312100/the Department of Science and Technology of Henan Province
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

