Human Respiratory Syncytial Virus Infection in a Human T Cell Line Is Hampered at Multiple Steps
- PMID: 33540662
- PMCID: PMC7913106
- DOI: 10.3390/v13020231
Human Respiratory Syncytial Virus Infection in a Human T Cell Line Is Hampered at Multiple Steps
Abstract
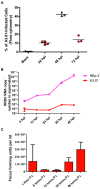
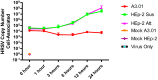
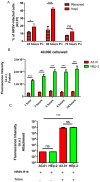
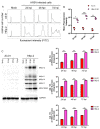
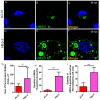
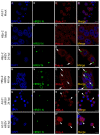
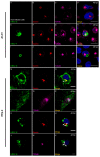
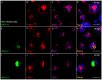
Human respiratory syncytial virus (HRSV) is the most frequent cause of severe respiratory disease in children. The main targets of HRSV infection are epithelial cells of the respiratory tract, and the great majority of the studies regarding HRSV infection are done in respiratory cells. Recently, the interest on respiratory virus infection of lymphoid cells has been growing, but details of the interaction of HRSV with lymphoid cells remain unknown. Therefore, this study was done to assess the relationship of HRSV with A3.01 cells, a human CD4+ T cell line. Using flow cytometry and fluorescent focus assay, we found that A3.01 cells are susceptible but virtually not permissive to HRSV infection. Dequenching experiments revealed that the fusion process of HRSV in A3.01 cells was nearly abolished in comparison to HEp-2 cells, an epithelial cell lineage. Quantification of viral RNA by RT-qPCR showed that the replication of HRSV in A3.01 cells was considerably reduced. Western blot and quantitative flow cytometry analyses demonstrated that the production of HRSV proteins in A3.01 was significantly lower than in HEp-2 cells. Additionally, using fluorescence in situ hybridization, we found that the inclusion body-associated granules (IBAGs) were almost absent in HRSV inclusion bodies in A3.01 cells. We also assessed the intracellular trafficking of HRSV proteins and found that HRSV proteins colocalized partially with the secretory pathway in A3.01 cells, but these HRSV proteins and viral filaments were present only scarcely at the plasma membrane. HRSV infection of A3.01 CD4+ T cells is virtually unproductive as compared to HEp-2 cells, as a result of defects at several steps of the viral cycle: Fusion, genome replication, formation of inclusion bodies, recruitment of cellular proteins, virus assembly, and budding.
Keywords: HRSV R18 fusion assay; HRSV entry; HRSV filament formation; HRSV inclusion body-associated granules (IBAG′s); HRSV intracellular trafficking; HRSV low protein production; human respiratory syncytial virus (HRSV) infection in T-cell line; inefficient HRSV replication A3.01.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
Similar articles
-
trans-Complementation allows recovery of human respiratory syncytial viruses that are infectious but deficient in cell-to-cell transmission.J Virol. 2004 Sep;78(17):9064-72. doi: 10.1128/JVI.78.17.9064-9072.2004. J Virol. 2004. PMID: 15308702 Free PMC article.
-
Modulation of Host Immunity by Human Respiratory Syncytial Virus Virulence Factors: A Synergic Inhibition of Both Innate and Adaptive Immunity.Front Cell Infect Microbiol. 2017 Aug 16;7:367. doi: 10.3389/fcimb.2017.00367. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28861397 Free PMC article. Review.
-
The Respiratory Syncytial Virus Phosphoprotein, Matrix Protein, and Fusion Protein Carboxy-Terminal Domain Drive Efficient Filamentous Virus-Like Particle Formation.J Virol. 2016 Nov 14;90(23):10612-10628. doi: 10.1128/JVI.01193-16. Print 2016 Dec 1. J Virol. 2016. PMID: 27654298 Free PMC article.
-
Use of a novel cell-based fusion reporter assay to explore the host range of human respiratory syncytial virus F protein.Virol J. 2005 Jul 13;2:54. doi: 10.1186/1743-422X-2-54. Virol J. 2005. PMID: 16014172 Free PMC article.
-
Contribution of Cytokines to Tissue Damage During Human Respiratory Syncytial Virus Infection.Front Immunol. 2019 Mar 18;10:452. doi: 10.3389/fimmu.2019.00452. eCollection 2019. Front Immunol. 2019. PMID: 30936869 Free PMC article. Review.
Cited by
-
Special Issue "Viral Infections in Developing Countries".Viruses. 2022 Feb 16;14(2):405. doi: 10.3390/v14020405. Viruses. 2022. PMID: 35215998 Free PMC article.
References
-
- Collins P.L., Karron R.A. Respiratory Syncytial Virus and Metapneumovirus. In: Knipe M.D., Howley M.P., editors. Fields Virology. 6th ed. Wolters Kluwer, Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2013. pp. 1086–1123.
-
- Modena J.L., Valera F.C., Jacob M.G., Buzatto G.P., Saturno T.H., Lopes L., Souza J.M., Paula F.E., Silva M.L., Carenzi L.R., et al. High rates of detection of respiratory viruses in tonsillar tissues from children with chronic adenotonsillar disease. PLoS ONE. 2012;7:e42136. doi: 10.1371/journal.pone.0042136. - DOI - PMC - PubMed
-
- Raiden S., Sananez I., Remes-Lenicov F., Pandolfi J., Romero C., De Lillo L., Ceballos A., Geffner J., Arruvito L. Respiratory syncytial virus (RSV) infects CD4+ T cells: Frequency of circulating CD4+ RSV+ T cells as a marker of disease severity in young children. J. Infect. Dis. 2017;215:1049–1058. doi: 10.1093/infdis/jix070. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

