Interplay between hevin, SPARC, and MDGAs: Modulators of neurexin-neuroligin transsynaptic bridges
- PMID: 33535026
- PMCID: PMC8254758
- DOI: 10.1016/j.str.2021.01.003
Interplay between hevin, SPARC, and MDGAs: Modulators of neurexin-neuroligin transsynaptic bridges
Abstract
Hevin is secreted by astrocytes and its synaptogenic effects are antagonized by the related protein, SPARC. Hevin stabilizes neurexin-neuroligin transsynaptic bridges in vivo. A third protein, membrane-tethered MDGA, blocks these bridges. Here, we reveal the molecular underpinnings of a regulatory network formed by this trio of proteins. The hevin FS-EC structure differs from SPARC, in that the EC domain appears rearranged around a conserved core. The FS domain is structurally conserved and it houses nanomolar affinity binding sites for neurexin and neuroligin. SPARC also binds neurexin and neuroligin, competing with hevin, so its antagonist action is rooted in its shortened N-terminal region. Strikingly, the hevin FS domain competes with MDGA for an overlapping binding site on neuroligin, while the hevin EC domain binds the extracellular matrix protein collagen (like SPARC), so that this trio of proteins can regulate neurexin-neuroligin transsynaptic bridges and also extracellular matrix interactions, impacting synapse formation and ultimately neural circuits.
Keywords: MDGAs; SPARC; adhesion molecule; hevin; matricellular protein; neurexins; neuroligins; neuropsychiatric disease; protein structure; synaptic organizer.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



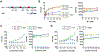


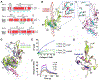
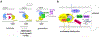
Comment in
-
Building and destroying synaptic bridges: How do Hevin/Sparcl1, SPARC, and MDGAs modify trans-synaptic neurexin-neuroligin interactions?Structure. 2021 Jul 1;29(7):635-637. doi: 10.1016/j.str.2021.06.011. Structure. 2021. PMID: 34214438
Similar articles
-
Control of excitatory CNS synaptogenesis by astrocyte-secreted proteins Hevin and SPARC.Proc Natl Acad Sci U S A. 2011 Aug 9;108(32):E440-9. doi: 10.1073/pnas.1104977108. Epub 2011 Jul 25. Proc Natl Acad Sci U S A. 2011. PMID: 21788491 Free PMC article.
-
Molecular Mechanism of MDGA1: Regulation of Neuroligin 2:Neurexin Trans-synaptic Bridges.Neuron. 2017 Jun 21;94(6):1132-1141.e4. doi: 10.1016/j.neuron.2017.06.009. Neuron. 2017. PMID: 28641112 Free PMC article.
-
Building and destroying synaptic bridges: How do Hevin/Sparcl1, SPARC, and MDGAs modify trans-synaptic neurexin-neuroligin interactions?Structure. 2021 Jul 1;29(7):635-637. doi: 10.1016/j.str.2021.06.011. Structure. 2021. PMID: 34214438
-
Hevin/SC1, a matricellular glycoprotein and potential tumor-suppressor of the SPARC/BM-40/Osteonectin family.Int J Biochem Cell Biol. 2004 Jun;36(6):991-6. doi: 10.1016/j.biocel.2004.01.017. Int J Biochem Cell Biol. 2004. PMID: 15094114 Review.
-
SPARC, a matricellular glycoprotein with important biological functions.J Histochem Cytochem. 1999 Dec;47(12):1495-506. doi: 10.1177/002215549904701201. J Histochem Cytochem. 1999. PMID: 10567433 Review.
Cited by
-
Autism-associated mutation in Hevin/Sparcl1 induces endoplasmic reticulum stress through structural instability.Sci Rep. 2022 Jul 13;12(1):11891. doi: 10.1038/s41598-022-15784-5. Sci Rep. 2022. PMID: 35831437 Free PMC article.
-
New perspective on sustained antidepressant effect: focus on neurexins regulating synaptic plasticity.Cell Death Discov. 2024 May 1;10(1):205. doi: 10.1038/s41420-024-01974-9. Cell Death Discov. 2024. PMID: 38693106 Free PMC article. Review.
-
MDGAs are fast-diffusing molecules that delay excitatory synapse development by altering neuroligin behavior.Elife. 2022 May 9;11:e75233. doi: 10.7554/eLife.75233. Elife. 2022. PMID: 35532105 Free PMC article.
-
Designer molecules of the synaptic organizer MDGA1 reveal 3D conformational control of biological function.J Biol Chem. 2023 Apr;299(4):104586. doi: 10.1016/j.jbc.2023.104586. Epub 2023 Mar 7. J Biol Chem. 2023. PMID: 36889589 Free PMC article.
-
Mutations in Hevin/Sparcl1 and risk of autism spectrum disorder.Neural Regen Res. 2023 Jul;18(7):1499-1500. doi: 10.4103/1673-5374.361543. Neural Regen Res. 2023. PMID: 36571352 Free PMC article. No abstract available.
References
-
- Albrecht D, López-Murcia FJ, Pérez-González AP, Lichtner G, Solsona C, and Llobet A (2012). SPARC prevents maturation of cholinergic presynaptic terminals. Mol. Cell. Neurosci 49, 364–374. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

