A Highly Contiguous Genome Assembly of a Polyphagous Predatory Mite Stratiolaelaps scimitus (Womersley) (Acari: Laelapidae)
- PMID: 33528489
- PMCID: PMC7936031
- DOI: 10.1093/gbe/evab011
A Highly Contiguous Genome Assembly of a Polyphagous Predatory Mite Stratiolaelaps scimitus (Womersley) (Acari: Laelapidae)
Abstract
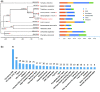
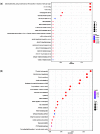
As a polyphagous soil-dwelling predatory mite, Stratiolaelaps scimitus (Womersley) (Acari: Laelapidae), formerly known as Stratiolaelaps miles (Berlese), is native to the Northern hemisphere and preys on soil invertebrates, including fungus gnats, springtails, thrips nymphs, nematodes, and other species of mites. Already mass-produced and commercialized in North America, Europe, Oceania and China, S. scimitus will highly likely be introduced to other countries and regions as a biocontrol agent against edaphic pests in the near future. The introduction, however, can lead to unexpected genetic changes within populations of biological control agents, which might decrease the efficacy of pest management or increase the risks to local environments. To better understand the genetic basis of its biology and behavior, we sequenced and assembled the draft genome of S. scimitus using the PacBio Sequel platform II. We generated ∼150× (64.81 Gb) PacBio long reads with an average read length of 12.60 kb. Reads longer than 5 kb were assembled into contigs, resulting in the final assembly of 158 contigs with an N50 length of 7.66 Mb, and captured 93.1% of the BUSCO (Benchmarking Universal Single-Copy Orthologs) gene set (n = 1,066). We identified 16.39% (69.91 Mb) repetitive elements, 1,686 noncoding RNAs, and 13,305 protein-coding genes, which represented 95.8% BUSCO completeness. Combining analyses of genome family evolution and function enrichment of gene ontology and pathway, a total of 135 families experienced significant expansions, which were mainly involved in digestion, detoxification, immunity, and venom. Major expansions of the detoxification enzymes, that is, P450s and carboxylesterases, suggest a possible genetic mechanism underlying polyphagy and ecological adaptions. Our high-quality genome assembly and annotation provide new insights on the evolutionary biology, soil ecology, and biological control for predaceous mites.
Keywords: Laelapidae; comparative genomics; detoxification; gene family evolution; genome annotation.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
Similar articles
-
Development and reproduction of Stratiolaelaps scimitus (Acari: Laelapidae) with fungus gnat larvae (Diptera: Sciaridae), potworms (Oligochaeta: Enchytraeidae) or Sancassania aff. sphaerogaster (Acari: Acaridae) as the sole food source.Exp Appl Acarol. 2005;36(1-2):71-81. doi: 10.1007/s10493-005-0242-x. Exp Appl Acarol. 2005. PMID: 16082925
-
Effects of greenhouse pesticides on the soil-dwelling predatory mite Stratiolaelaps scimitus (Acari: Mesostigmata: Laelapidae) under laboratory conditions.J Econ Entomol. 2004 Jun;97(3):793-9. doi: 10.1093/jee/97.3.793. J Econ Entomol. 2004. PMID: 15279255
-
Potential of astigmatid mites (Acari: Astigmatina) as prey for rearing edaphic predatory mites of the families Laelapidae and Rhodacaridae (Acari: Mesostigmata).Exp Appl Acarol. 2016 Jul;69(3):289-96. doi: 10.1007/s10493-016-0043-4. Epub 2016 Apr 26. Exp Appl Acarol. 2016. PMID: 27115501
-
Compatibility of pesticides used in strawberry crops with predatory mites Stratiolaelaps scimitus (Womersley) and Cosmolaelaps brevistilis (Karg).Ecotoxicology. 2020 Mar;29(2):148-155. doi: 10.1007/s10646-020-02164-w. Epub 2020 Jan 11. Ecotoxicology. 2020. PMID: 31927677
-
Whole genome sequencing and annotation of Scleroderma yunnanense, the only edible Scleroderma species.Genomics. 2023 Nov;115(6):110727. doi: 10.1016/j.ygeno.2023.110727. Epub 2023 Oct 14. Genomics. 2023. PMID: 37839651 Review.
Cited by
-
A Long-Read Genome Assembly of a Native Mite in China Pyemotes zhonghuajia Yu, Zhang & He (Prostigmata: Pyemotidae) Reveals Gene Expansion in Toxin-Related Gene Families.Toxins (Basel). 2022 Aug 21;14(8):571. doi: 10.3390/toxins14080571. Toxins (Basel). 2022. PMID: 36006233 Free PMC article.
-
Molecular phylogeny reveals Varroa mites are not a separate family but a subfamily of Laelapidae.Sci Rep. 2024 Jun 18;14(1):13994. doi: 10.1038/s41598-024-63991-z. Sci Rep. 2024. PMID: 38886408 Free PMC article.
References
-
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12 (1):59–60. - PubMed
-
- Bushnell B. 2014. BBtools. Available from: https://sourceforge.net/projects/bbmap/.
-
- Cabrera AR, Cloyd RA, Zaborski ER.. 2005. Development and reproduction of Stratiolaelaps scimitus (Acari: laelapidae) with fungus gnat larvae (Diptera: sciaridae), potworms (Oligochaeta: enchytraeidae) or Sancassaniaaff Sphaerogaster (Acari: acaridae) as the sole food source. Exp Appl Acarol. 36(1–2):71–81. - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

