Cell Cycle Regulation of the Pdx1 Transcription Factor in Developing Pancreas and Insulin-Producing β-Cells
- PMID: 33526589
- PMCID: PMC7980191
- DOI: 10.2337/db20-0599
Cell Cycle Regulation of the Pdx1 Transcription Factor in Developing Pancreas and Insulin-Producing β-Cells
Abstract
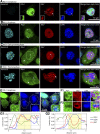
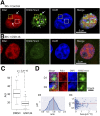
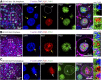
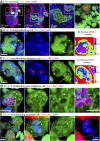
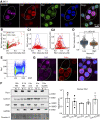
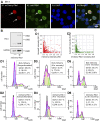
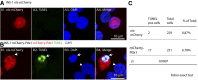
Current evidence indicates that proliferating β-cells express lower levels of some functional cell identity genes, suggesting that proliferating cells are not optimally functional. Pdx1 is important for β-cell specification, function, and proliferation and is mutated in monogenic forms of diabetes. However, its regulation during the cell cycle is unknown. Here we examined Pdx1 protein expression in immortalized β-cells, maternal mouse islets during pregnancy, and mouse embryonic pancreas. We demonstrate that Pdx1 localization and protein levels are highly dynamic. In nonmitotic cells, Pdx1 is not observed in constitutive heterochromatin, nucleoli, or most areas containing repressive epigenetic marks. At prophase, Pdx1 is enriched around the chromosomes before Ki67 coating of the chromosome surface. Pdx1 uniformly localizes in the cytoplasm at prometaphase and becomes enriched around the chromosomes again at the end of cell division, before nuclear envelope formation. Cells in S phase have lower Pdx1 levels than cells at earlier cell cycle stages, and overexpression of Pdx1 in INS-1 cells prevents progression toward G2, suggesting that cell cycle-dependent regulation of Pdx1 is required for completion of mitosis. Together, we find that Pdx1 localization and protein levels are tightly regulated throughout the cell cycle. This dynamic regulation has implications for the dichotomous role of Pdx1 in β-cell function and proliferation.
© 2021 by the American Diabetes Association.
Figures
Similar articles
-
Biphasic induction of Pdx1 in mouse and human embryonic stem cells can mimic development of pancreatic beta-cells.Stem Cells. 2009 Feb;27(2):341-51. doi: 10.1634/stemcells.2008-0310. Stem Cells. 2009. PMID: 19056911
-
Defining a Novel Role for the Pdx1 Transcription Factor in Islet β-Cell Maturation and Proliferation During Weaning.Diabetes. 2017 Nov;66(11):2830-2839. doi: 10.2337/db16-1516. Epub 2017 Jul 13. Diabetes. 2017. PMID: 28705881 Free PMC article.
-
Differentiation of Mouse Embryonic Stem Cells toward Functional Pancreatic β-Cell Surrogates through Epigenetic Regulation of Pdx1 by Nitric Oxide.Cell Transplant. 2016 Oct;25(10):1879-1892. doi: 10.3727/096368916X691178. Cell Transplant. 2016. PMID: 26980118
-
PDX1, Neurogenin-3, and MAFA: critical transcription regulators for beta cell development and regeneration.Stem Cell Res Ther. 2017 Nov 2;8(1):240. doi: 10.1186/s13287-017-0694-z. Stem Cell Res Ther. 2017. PMID: 29096722 Free PMC article. Review.
-
Impact of Pdx1-associated chromatin modifiers on islet β-cells.Diabetes Obes Metab. 2016 Sep;18 Suppl 1(Suppl 1):123-7. doi: 10.1111/dom.12730. Diabetes Obes Metab. 2016. PMID: 27615141 Free PMC article. Review.
Cited by
-
Txnip deficiency promotes β-cell proliferation in the HFD-induced obesity mouse model.Endocr Connect. 2022 Apr 29;11(4):e210641. doi: 10.1530/EC-21-0641. Endocr Connect. 2022. PMID: 35294398 Free PMC article.
-
Regulation of Pdx1 by oxidative stress and Nrf2 in pancreatic beta-cells.Front Endocrinol (Lausanne). 2022 Sep 15;13:1011187. doi: 10.3389/fendo.2022.1011187. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36187092 Free PMC article. Review.
-
Molecular Mechanism of Pancreatic β-Cell Failure in Type 2 Diabetes Mellitus.Biomedicines. 2022 Mar 31;10(4):818. doi: 10.3390/biomedicines10040818. Biomedicines. 2022. PMID: 35453568 Free PMC article. Review.
-
Targeting pancreatic beta cell death in type 2 diabetes by polyphenols.Front Endocrinol (Lausanne). 2022 Nov 17;13:1052317. doi: 10.3389/fendo.2022.1052317. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36465657 Free PMC article. Review.
-
Characterization and functional evaluation of goat PDX1 regulatory modules through comparative analysis of conserved interspecies homologs.Sci Rep. 2024 Nov 5;14(1):26755. doi: 10.1038/s41598-024-77614-0. Sci Rep. 2024. PMID: 39500950 Free PMC article.
References
-
- Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature 2004;429:41–46 - PubMed
-
- Fischer M, Müller GA. Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes. Crit Rev Biochem Mol Biol 2017;52:638–662 - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

