Expression of Endogenous Angiotensin-Converting Enzyme 2 in Human Induced Pluripotent Stem Cell-Derived Retinal Organoids
- PMID: 33525682
- PMCID: PMC7865454
- DOI: 10.3390/ijms22031320
Expression of Endogenous Angiotensin-Converting Enzyme 2 in Human Induced Pluripotent Stem Cell-Derived Retinal Organoids
Abstract
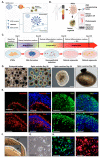
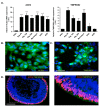
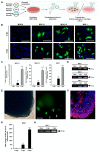
Angiotensin-converting enzyme 2 (ACE2) was identified as the main host cell receptor for the entry of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its subsequent infection. In some coronavirus disease 2019 (COVID-19) patients, it has been reported that the nervous tissues and the eyes were also affected. However, evidence supporting that the retina is a target tissue for SARS-CoV-2 infection is still lacking. This present study aimed to investigate whether ACE2 expression plays a role in human retinal neurons during SARS-CoV-2 infection. Human induced pluripotent stem cell (hiPSC)-derived retinal organoids and monolayer cultures derived from dissociated retinal organoids were generated. To validate the potential entry of SARS-CoV-2 infection in the retina, we showed that hiPSC-derived retinal organoids and monolayer cultures endogenously express ACE2 and transmembrane serine protease 2 (TMPRSS2) on the mRNA level. Immunofluorescence staining confirmed the protein expression of ACE2 and TMPRSS2 in retinal organoids and monolayer cultures. Furthermore, using the SARS-CoV-2 pseudovirus spike protein with GFP expression system, we found that retinal organoids and monolayer cultures can potentially be infected by the SARS-CoV-2 pseudovirus. Collectively, our findings highlighted the potential of iPSC-derived retinal organoids as the models for ACE2 receptor-based SARS-CoV-2 infection.
Keywords: ACE2; COVID-19; SARS-CoV-2; SARS-CoV-2 pseudovirus; induced pluripotent stem cells; organoids; spike protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
SARS-CoV-2 and SARS-CoV Spike-Mediated Cell-Cell Fusion Differ in Their Requirements for Receptor Expression and Proteolytic Activation.J Virol. 2021 Apr 12;95(9):e00002-21. doi: 10.1128/JVI.00002-21. Print 2021 Apr 12. J Virol. 2021. PMID: 33608407 Free PMC article.
-
SARS-CoV-2 pseudovirus infectivity and expression of viral entry-related factors ACE2, TMPRSS2, Kim-1, and NRP-1 in human cells from the respiratory, urinary, digestive, reproductive, and immune systems.J Med Virol. 2021 Dec;93(12):6671-6685. doi: 10.1002/jmv.27244. Epub 2021 Aug 4. J Med Virol. 2021. PMID: 34324210 Free PMC article.
-
Contributions of human ACE2 and TMPRSS2 in determining host-pathogen interaction of COVID-19.J Genet. 2021;100(1):12. doi: 10.1007/s12041-021-01262-w. J Genet. 2021. PMID: 33707363 Free PMC article. Review.
-
Kidney organoids reveal redundancy in viral entry pathways during ACE2-dependent SARS-CoV-2 infection.J Virol. 2024 Mar 19;98(3):e0180223. doi: 10.1128/jvi.01802-23. Epub 2024 Feb 9. J Virol. 2024. PMID: 38334329 Free PMC article.
-
Targeting the viral-entry facilitators of SARS-CoV-2 as a therapeutic strategy in COVID-19.J Med Virol. 2021 Sep;93(9):5260-5276. doi: 10.1002/jmv.27019. Epub 2021 May 3. J Med Virol. 2021. PMID: 33851732 Free PMC article. Review.
Cited by
-
MSCs vs. iPSCs: Potential in therapeutic applications.Front Cell Dev Biol. 2022 Nov 2;10:1005926. doi: 10.3389/fcell.2022.1005926. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36407112 Free PMC article. Review.
-
Human organoid models to study SARS-CoV-2 infection.Nat Methods. 2022 Apr;19(4):418-428. doi: 10.1038/s41592-022-01453-y. Epub 2022 Apr 8. Nat Methods. 2022. PMID: 35396481 Review.
-
Using 2D and 3D pluripotent stem cell models to study neurotropic viruses.Front Virol. 2022;2:869657. doi: 10.3389/fviro.2022.869657. Epub 2022 Jul 29. Front Virol. 2022. PMID: 36325520 Free PMC article.
-
Exploring the promising potential of induced pluripotent stem cells in cancer research and therapy.Mol Cancer. 2023 Nov 28;22(1):189. doi: 10.1186/s12943-023-01873-0. Mol Cancer. 2023. PMID: 38017433 Free PMC article. Review.
-
The Role of Small Molecules and Their Effect on the Molecular Mechanisms of Early Retinal Organoid Development.Int J Mol Sci. 2021 Jun 30;22(13):7081. doi: 10.3390/ijms22137081. Int J Mol Sci. 2021. PMID: 34209272 Free PMC article. Review.
References
-
- Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. Features, Evaluation, and Treatment of Coronavirus (COVID-19) StatPearls; Treasure Island, FL, USA: 2020. - PubMed
-
- Schmitt C.A., Bergey C.M., Jasinska A.J., Ramensky V., Burt F., Svardal H., Jorgensen M.J., Freimer N.B., Grobler J.P., Turner T.R. ACE2 and TMPRSS2 variation in savanna monkeys (Chlorocebus spp.): Potential risk for zoonotic/anthroponotic transmission of SARS-CoV-2 and a potential model for functional studies. PLoS ONE. 2020;15:e0235106. doi: 10.1371/journal.pone.0235106. - DOI - PMC - PubMed
-
- Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffre F., et al. A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell. 2020;27:125–136.e7. doi: 10.1016/j.stem.2020.06.015. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

