Intake of Natural Compounds and Circulating microRNA Expression Levels: Their Relationship Investigated in Healthy Subjects With Different Dietary Habits
- PMID: 33519486
- PMCID: PMC7840481
- DOI: 10.3389/fphar.2020.619200
Intake of Natural Compounds and Circulating microRNA Expression Levels: Their Relationship Investigated in Healthy Subjects With Different Dietary Habits
Abstract
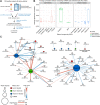
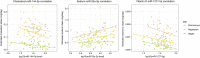
Diet has a strong influence on many physiological processes, which in turn have important implications on a variety of pathological conditions. In this respect, microRNAs (miRNAs), a class of small non-coding RNAs playing a relevant epigenetic role in controlling gene expression, may represent mediators between the dietary intake and the healthy status. Despite great advances in the field of nutri-epigenomics, it remains unclear how miRNA expression is modulated by the diet and, specifically, the intake of specific nutrients. We investigated the whole circulating miRNome by small RNA-sequencing performed on plasma samples of 120 healthy volunteers with different dietary habits (vegans, vegetarians, and omnivores). Dietary intakes of specific nutrients were estimated for each subject from the information reported in the food-frequency questionnaire previously validated in the EPIC study. We focused hereby on the intake of 23 natural compounds (NCs) of the classes of lipids, micro-elements, and vitamins. We identified 78 significant correlations (rho > 0.300, p-value < 0.05) among the estimated daily intake of 13 NCs and the expression levels of 58 plasma miRNAs. Overall, vitamin D, sodium, and vitamin E correlated with the largest number of miRNAs. All the identified correlations were consistent among the three dietary groups and 22 of them were confirmed as significant (p-value < 0.05) by age-, gender-, and body-mass index-adjusted Generalized Linear regression Model analysis. miR-23a-3p expression levels were related with different NCs including a significant positive correlation with sodium (rho = 0.377) and significant negative correlations with lipid-related NCs and vitamin E. Conversely, the estimated intake of vitamin D was negatively correlated with the expression of the highest number of circulating miRNAs, particularly miR-1277-5p (rho = -0.393) and miR-144-3p (rho = -0.393). Functional analysis of the targets of sodium intake-correlated miRNAs highlighted terms related to cardiac development. A similar approach on targets of those miRNAs correlated with vitamin D intake showed an enrichment in genes involved in hormone metabolisms, while the response to chronic inflammation was among the top enriched processes involving targets of miRNAs negatively related with vitamin E intake. Our findings show that nutrients through the habitual diet influence circulating miRNA profiles and highlight that this aspect must be considered in the nutri-epigenomic research.
Keywords: circulating miRNA; dietary habits; dietary natural compounds; plasma metabolites; sodium; vitamin D; vitamin E.
Copyright © 2021 Ferrero, Carpi, Polini, Pardini, Nieri, Impeduglia, Grioni, Tarallo and Naccarati.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Stool microRNA profiles reflect different dietary and gut microbiome patterns in healthy individuals.Gut. 2022 Jul;71(7):1302-1314. doi: 10.1136/gutjnl-2021-325168. Epub 2021 Jul 27. Gut. 2022. PMID: 34315772 Free PMC article.
-
MicroRNA expression in relation to different dietary habits: a comparison in stool and plasma samples.Mutagenesis. 2014 Sep;29(5):385-91. doi: 10.1093/mutage/geu028. Mutagenesis. 2014. PMID: 25150024
-
Dysregulation of endothelial colony-forming cell function by a negative feedback loop of circulating miR-146a and -146b in cardiovascular disease patients.PLoS One. 2017 Jul 20;12(7):e0181562. doi: 10.1371/journal.pone.0181562. eCollection 2017. PLoS One. 2017. PMID: 28727754 Free PMC article.
-
Diet and obstructive lung diseases.Epidemiol Rev. 2001;23(2):268-87. doi: 10.1093/oxfordjournals.epirev.a000806. Epidemiol Rev. 2001. PMID: 12192737 Review.
-
The Impact of Diet on miRNA Regulation and Its Implications for Health: A Systematic Review.Nutrients. 2024 Mar 7;16(6):770. doi: 10.3390/nu16060770. Nutrients. 2024. PMID: 38542682 Free PMC article. Review.
Cited by
-
Exerkines, Nutrition, and Systemic Metabolism.Nutrients. 2024 Jan 30;16(3):410. doi: 10.3390/nu16030410. Nutrients. 2024. PMID: 38337694 Free PMC article. Review.
-
Stool microRNA profiles reflect different dietary and gut microbiome patterns in healthy individuals.Gut. 2022 Jul;71(7):1302-1314. doi: 10.1136/gutjnl-2021-325168. Epub 2021 Jul 27. Gut. 2022. PMID: 34315772 Free PMC article.
-
Specific circulating miRNAs are associated with plasma lipids in a healthy American cohort.Physiol Genomics. 2024 Jul 1;56(7):492-505. doi: 10.1152/physiolgenomics.00087.2023. Epub 2024 Apr 1. Physiol Genomics. 2024. PMID: 38557280
-
Tanshinone IIA and Cryptotanshinone Counteract Inflammation by Regulating Gene and miRNA Expression in Human SGBS Adipocytes.Biomolecules. 2023 Jun 23;13(7):1029. doi: 10.3390/biom13071029. Biomolecules. 2023. PMID: 37509065 Free PMC article.
-
Circulating non-coding RNA biomarkers of endocrine tumours.Nat Rev Endocrinol. 2024 Oct;20(10):600-614. doi: 10.1038/s41574-024-01005-8. Epub 2024 Jun 17. Nat Rev Endocrinol. 2024. PMID: 38886617 Review.
References
-
- Beckett E. L., Martin C., Duesing K., Jones P., Furst J., Yates Z., et al. (2014). Vitamin D receptor genotype modulates the correlation between vitamin D and circulating levels of let-7a/b and vitamin D intake in an elderly cohort. J. Nutrigenet. Nutrigenomics 7, 264–273. 10.1159/000381676 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

