Complexity in genetic cardiomyopathies and new approaches for mechanism-based precision medicine
- PMID: 33512404
- PMCID: PMC7852459
- DOI: 10.1085/jgp.202012662
Complexity in genetic cardiomyopathies and new approaches for mechanism-based precision medicine
Abstract
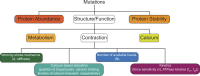
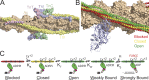
Genetic cardiomyopathies have been studied for decades, and it has become increasingly clear that these progressive diseases are more complex than originally thought. These complexities can be seen both in the molecular etiologies of these disorders and in the clinical phenotypes observed in patients. While these disorders can be caused by mutations in cardiac genes, including ones encoding sarcomeric proteins, the disease presentation varies depending on the patient mutation, where mutations even within the same gene can cause divergent phenotypes. Moreover, it is challenging to connect the mutation-induced molecular insult that drives the disease pathogenesis with the various compensatory and maladaptive pathways that are activated during the course of the subsequent progressive, pathogenic cardiac remodeling. These inherent complexities have frustrated our ability to understand and develop broadly effective treatments for these disorders. It has been proposed that it might be possible to improve patient outcomes by adopting a precision medicine approach. Here, we lay out a practical framework for such an approach, where patient subpopulations are binned based on common underlying biophysical mechanisms that drive the molecular disease pathogenesis, and we propose that this function-based approach will enable the development of targeted therapeutics that ameliorate these effects. We highlight several mutations to illustrate the need for mechanistic molecular experiments that span organizational and temporal scales, and we describe recent advances in the development of novel therapeutics based on functional targets. Finally, we describe many of the outstanding questions for the field and how fundamental mechanistic studies, informed by our more nuanced understanding of the clinical disorders, will play a central role in realizing the potential of precision medicine for genetic cardiomyopathies.
© 2021 Greenberg and Tardiff.
Figures


Similar articles
-
Sarcomeric proteins and inherited cardiomyopathies.Cardiovasc Res. 2008 Mar 1;77(4):659-66. doi: 10.1093/cvr/cvm084. Epub 2007 Dec 4. Cardiovasc Res. 2008. PMID: 18056765 Review.
-
Moving beyond simple answers to complex disorders in sarcomeric cardiomyopathies: the role of integrated systems.Pflugers Arch. 2019 May;471(5):661-671. doi: 10.1007/s00424-019-02269-0. Epub 2019 Mar 8. Pflugers Arch. 2019. PMID: 30848350 Free PMC article. Review.
-
Contemporary and Future Approaches to Precision Medicine in Inherited Cardiomyopathies: JACC Focus Seminar 3/5.J Am Coll Cardiol. 2021 May 25;77(20):2551-2572. doi: 10.1016/j.jacc.2020.12.072. J Am Coll Cardiol. 2021. PMID: 34016267 Review.
-
Untying the knot: protein quality control in inherited cardiomyopathies.Pflugers Arch. 2019 May;471(5):795-806. doi: 10.1007/s00424-018-2194-0. Epub 2018 Aug 14. Pflugers Arch. 2019. PMID: 30109411 Free PMC article. Review.
-
Genetic advances in sarcomeric cardiomyopathies: state of the art.Cardiovasc Res. 2015 Apr 1;105(4):397-408. doi: 10.1093/cvr/cvv025. Epub 2015 Jan 29. Cardiovasc Res. 2015. PMID: 25634555 Free PMC article. Review.
Cited by
-
Insights into the Mechanism of the Cardiac Drug Omecamtiv Mecarbil─A Computational Study.J Phys Chem B. 2022 Dec 8;126(48):10069-10082. doi: 10.1021/acs.jpcb.2c06679. Epub 2022 Nov 29. J Phys Chem B. 2022. PMID: 36448224 Free PMC article.
-
Translation of New and Emerging Therapies for Genetic Cardiomyopathies.JACC Basic Transl Sci. 2021 Dec 1;7(1):70-83. doi: 10.1016/j.jacbts.2021.07.012. eCollection 2022 Jan. JACC Basic Transl Sci. 2021. PMID: 35128211 Free PMC article. Review.
-
Mechanical dysfunction of the sarcomere induced by a pathogenic mutation in troponin T drives cellular adaptation.J Gen Physiol. 2021 May 3;153(5):e202012787. doi: 10.1085/jgp.202012787. J Gen Physiol. 2021. PMID: 33856419 Free PMC article.
-
Drug specificity and affinity are encoded in the probability of cryptic pocket opening in myosin motor domains.Elife. 2023 Jan 27;12:e83602. doi: 10.7554/eLife.83602. Elife. 2023. PMID: 36705568 Free PMC article.
-
Excitation-contraction coupling in cardiac, skeletal, and smooth muscle.J Gen Physiol. 2022 Sep 5;154(9):e202213244. doi: 10.1085/jgp.202213244. Epub 2022 Aug 19. J Gen Physiol. 2022. PMID: 35984377 Free PMC article. No abstract available.
References
-
- Adhikari, A.S., Trivedi D.V., Sarkar S.S., Song D., Kooiker K.B., Bernstein D., Spudich J.A., and Ruppel K.M.. 2019. β-Cardiac myosin hypertrophic cardiomyopathy mutations release sequestered heads and increase enzymatic activity. Nat. Commun. 10:2685. 10.1038/s41467-019-10555-9 - DOI - PMC - PubMed
-
- Ahmad, F., Banerjee S.K., Lage M.L., Huang X.N., Smith S.H., Saba S., Rager J., Conner D.A., Janczewski A.M., Tobita K., et al. . 2008. The role of cardiac troponin T quantity and function in cardiac development and dilated cardiomyopathy. PLoS One. 3:e2642. 10.1371/journal.pone.0002642 - DOI - PMC - PubMed
-
- Alamo, L., Qi D., Wriggers W., Pinto A., Zhu J., Bilbao A., Gillilan R.E., Hu S., and Padrón R.. 2016. Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis. J. Mol. Biol. 428:1142–1164. 10.1016/j.jmb.2016.01.027 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

