Arabidopsis REI-LIKE proteins activate ribosome biogenesis during cold acclimation
- PMID: 33510206
- PMCID: PMC7844247
- DOI: 10.1038/s41598-021-81610-z
Arabidopsis REI-LIKE proteins activate ribosome biogenesis during cold acclimation
Abstract
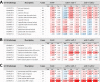
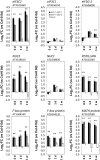
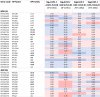
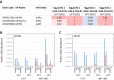
Arabidopsis REIL proteins are cytosolic ribosomal 60S-biogenesis factors. After shift to 10 °C, reil mutants deplete and slowly replenish non-translating eukaryotic ribosome complexes of root tissue, while controlling the balance of non-translating 40S- and 60S-subunits. Reil mutations respond by hyper-accumulation of non-translating subunits at steady-state temperature; after cold-shift, a KCl-sensitive 80S sub-fraction remains depleted. We infer that Arabidopsis may buffer fluctuating translation by pre-existing non-translating ribosomes before de novo synthesis meets temperature-induced demands. Reil1 reil2 double mutants accumulate 43S-preinitiation and pre-60S-maturation complexes and alter paralog composition of ribosomal proteins in non-translating complexes. With few exceptions, e.g. RPL3B and RPL24C, these changes are not under transcriptional control. Our study suggests requirement of de novo synthesis of eukaryotic ribosomes for long-term cold acclimation, feedback control of NUC2 and eIF3C2 transcription and links new proteins, AT1G03250, AT5G60530, to plant ribosome biogenesis. We propose that Arabidopsis requires biosynthesis of specialized ribosomes for cold acclimation.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Plant Temperature Acclimation and Growth Rely on Cytosolic Ribosome Biogenesis Factor Homologs.Plant Physiol. 2018 Mar;176(3):2251-2276. doi: 10.1104/pp.17.01448. Epub 2018 Jan 30. Plant Physiol. 2018. PMID: 29382692 Free PMC article.
-
Spatially Enriched Paralog Rearrangements Argue Functionally Diverse Ribosomes Arise during Cold Acclimation in Arabidopsis.Int J Mol Sci. 2021 Jun 7;22(11):6160. doi: 10.3390/ijms22116160. Int J Mol Sci. 2021. PMID: 34200446 Free PMC article.
-
The REIL1 and REIL2 proteins of Arabidopsis thaliana are required for leaf growth in the cold.Plant Physiol. 2013 Dec;163(4):1623-39. doi: 10.1104/pp.113.223925. Epub 2013 Sep 13. Plant Physiol. 2013. PMID: 24038679 Free PMC article.
-
Cold acclimation by the CBF-COR pathway in a changing climate: Lessons from Arabidopsis thaliana.Plant Cell Rep. 2019 May;38(5):511-519. doi: 10.1007/s00299-019-02376-3. Epub 2019 Jan 16. Plant Cell Rep. 2019. PMID: 30652229 Free PMC article. Review.
-
Gene Regulatory Networks Mediating Cold Acclimation: The CBF Pathway.Adv Exp Med Biol. 2018;1081:3-22. doi: 10.1007/978-981-13-1244-1_1. Adv Exp Med Biol. 2018. PMID: 30288701 Review.
Cited by
-
Genetic Architecture Underlying the Metabolites of Chlorogenic Acid Biosynthesis in Populus tomentosa.Int J Mol Sci. 2021 Feb 27;22(5):2386. doi: 10.3390/ijms22052386. Int J Mol Sci. 2021. PMID: 33673666 Free PMC article.
-
Regulatory dynamics of gene expression in the developing male gametophyte of Arabidopsis.Plant Reprod. 2023 Sep;36(3):213-241. doi: 10.1007/s00497-022-00452-5. Epub 2022 Oct 25. Plant Reprod. 2023. PMID: 36282332 Free PMC article.
-
Cryo-EM structure and rRNA modification sites of a plant ribosome.Plant Commun. 2022 Sep 12;3(5):100342. doi: 10.1016/j.xplc.2022.100342. Epub 2022 May 27. Plant Commun. 2022. PMID: 35643637 Free PMC article.
-
Dynamic Changes in Seed Germination under Low-Temperature Stress in Maize.Int J Mol Sci. 2022 May 14;23(10):5495. doi: 10.3390/ijms23105495. Int J Mol Sci. 2022. PMID: 35628306 Free PMC article.
-
Engineering Ribosomes to Alleviate Abiotic Stress in Plants: A Perspective.Plants (Basel). 2022 Aug 12;11(16):2097. doi: 10.3390/plants11162097. Plants (Basel). 2022. PMID: 36015400 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

