Induction of Native c-di-GMP Phosphodiesterases Leads to Dispersal of Pseudomonas aeruginosa Biofilms
- PMID: 33495218
- PMCID: PMC8097489
- DOI: 10.1128/AAC.02431-20
Induction of Native c-di-GMP Phosphodiesterases Leads to Dispersal of Pseudomonas aeruginosa Biofilms
Abstract
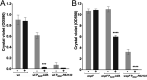
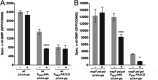
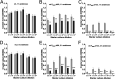
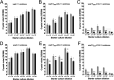
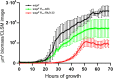
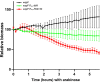
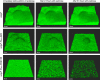
A decade of research has shown that the molecule c-di-GMP functions as a central second messenger in many bacteria. A high level of c-di-GMP is associated with biofilm formation, whereas a low level of c-di-GMP is associated with a planktonic single-cell bacterial lifestyle. c-di-GMP is formed by diguanylate cyclases and is degraded by specific phosphodiesterases. We previously presented evidence that the ectopic expression of the Escherichia coli phosphodiesterase YhjH in Pseudomonas aeruginosa results in biofilm dispersal. More recently, however, evidence has been presented that the induction of native c-di-GMP phosphodiesterases does not lead to a dispersal of P. aeruginosa biofilms. The latter result may discourage attempts to use c-di-GMP signaling as a target for the development of antibiofilm drugs. However, here, we demonstrate that the induction of the P. aeruginosa c-di-GMP phosphodiesterases PA2133 and BifA indeed results in the dispersal of P. aeruginosa biofilms in both a microtiter tray biofilm assay and a flow cell biofilm system.
Keywords: Pseudomonas aeruginosa; biofilm dispersal; c-di-GMP; phosphodiesterase.
Copyright © 2021 American Society for Microbiology.
Figures
Similar articles
-
Diguanylate Cyclases and Phosphodiesterases Required for Basal-Level c-di-GMP in Pseudomonas aeruginosa as Revealed by Systematic Phylogenetic and Transcriptomic Analyses.Appl Environ Microbiol. 2019 Oct 16;85(21):e01194-19. doi: 10.1128/AEM.01194-19. Print 2019 Nov 1. Appl Environ Microbiol. 2019. PMID: 31444209 Free PMC article.
-
Binding of GTP to BifA is required for the production of Pel-dependent biofilms in Pseudomonas aeruginosa.J Bacteriol. 2024 Feb 22;206(2):e0033123. doi: 10.1128/jb.00331-23. Epub 2024 Jan 10. J Bacteriol. 2024. PMID: 38197635 Free PMC article.
-
Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa.Proc Natl Acad Sci U S A. 2015 Sep 8;112(36):11359-64. doi: 10.1073/pnas.1421450112. Epub 2015 Aug 24. Proc Natl Acad Sci U S A. 2015. PMID: 26305928 Free PMC article.
-
c-di-GMP signaling in Pseudomonas syringae complex.Microbiol Res. 2023 Oct;275:127445. doi: 10.1016/j.micres.2023.127445. Epub 2023 Jul 6. Microbiol Res. 2023. PMID: 37450986 Review.
-
Molecular and structural facets of c-di-GMP signalling associated with biofilm formation in Pseudomonas aeruginosa.Mol Aspects Med. 2021 Oct;81:101001. doi: 10.1016/j.mam.2021.101001. Epub 2021 Jul 24. Mol Aspects Med. 2021. PMID: 34311995 Review.
Cited by
-
The dynamics of biofilm development and dispersal should be taken into account when quantifying biofilm via the crystal violet microtiter plate assay.Biofilm. 2024 Jun 20;8:100207. doi: 10.1016/j.bioflm.2024.100207. eCollection 2024 Dec. Biofilm. 2024. PMID: 39021701 Free PMC article.
-
Controlling Biofilm Development Through Cyclic di-GMP Signaling.Adv Exp Med Biol. 2022;1386:69-94. doi: 10.1007/978-3-031-08491-1_3. Adv Exp Med Biol. 2022. PMID: 36258069 Free PMC article. Review.
-
Inhibition of Pseudomonas aeruginosa quorum sensing by chemical induction of the MexEF-oprN efflux pump.Antimicrob Agents Chemother. 2024 Feb 7;68(2):e0138723. doi: 10.1128/aac.01387-23. Epub 2024 Jan 8. Antimicrob Agents Chemother. 2024. PMID: 38189278 Free PMC article.
-
The Alginate and Motility Regulator AmrZ is Essential for the Regulation of the Dispersion Response by Pseudomonas aeruginosa Biofilms.mSphere. 2022 Dec 21;7(6):e0050522. doi: 10.1128/msphere.00505-22. Epub 2022 Nov 14. mSphere. 2022. PMID: 36374041 Free PMC article.
-
What's in a name? Characteristics of clinical biofilms.FEMS Microbiol Rev. 2023 Sep 5;47(5):fuad050. doi: 10.1093/femsre/fuad050. FEMS Microbiol Rev. 2023. PMID: 37656883 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

