Kidney injury molecule-1 is a potential receptor for SARS-CoV-2
- PMID: 33493263
- PMCID: PMC7928767
- DOI: 10.1093/jmcb/mjab003
Kidney injury molecule-1 is a potential receptor for SARS-CoV-2
Abstract
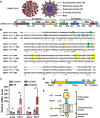
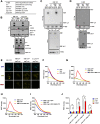
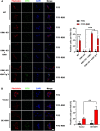
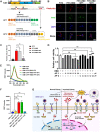
COVID-19 patients present high incidence of kidney abnormalities, which are associated with poor prognosis and mortality. The identification of SARS-CoV-2 in the kidney of COVID-19 patients suggests renal tropism of SARS-CoV-2. However, whether there is a specific target of SARS-CoV-2 in the kidney remains unclear. Herein, by using in silico simulation, coimmunoprecipitation, fluorescence resonance energy transfer, fluorescein isothiocyanate labeling, and rational design of antagonist peptides, we demonstrate that kidney injury molecule-1 (KIM1), a molecule dramatically upregulated upon kidney injury, binds with the receptor-binding domain (RBD) of SARS-CoV-2 and facilitates its attachment to cell membrane, with the immunoglobulin variable Ig-like (Ig V) domain of KIM1 playing a key role in this recognition. The interaction between SARS-CoV-2 RBD and KIM1 is potently blockaded by a rationally designed KIM1-derived polypeptide AP2. In addition, our results also suggest interactions between KIM1 Ig V domain and the RBDs of SARS-CoV and MERS-CoV, pathogens of two severe infectious respiratory diseases. Together, these findings suggest KIM1 as a novel receptor for SARS-CoV-2 and other coronaviruses. We propose that KIM1 may thus mediate and exacerbate the renal infection of SARS-CoV-2 in a 'vicious cycle', and KIM1 could be further explored as a therapeutic target.
Keywords: COVID-19; SARS-CoV-2; coronavirus; kidney diseases; kidney injury molecule-1.
© The Author(s) (2021). Published by Oxford University Press on behalf of Journal of Molecular Cell Biology, CEMCS, CAS.
Figures

Similar articles
-
Kidney injury molecule-1: a novel entry factor for SARS-CoV-2.J Mol Cell Biol. 2021 Jul 6;13(3):159-160. doi: 10.1093/jmcb/mjab006. J Mol Cell Biol. 2021. PMID: 33453103 Free PMC article. No abstract available.
-
Identification of a special cell type as a determinant of the kidney tropism of SARS-CoV-2.FEBS J. 2021 Sep;288(17):5163-5178. doi: 10.1111/febs.16114. Epub 2021 Jul 29. FEBS J. 2021. PMID: 34228902 Free PMC article.
-
Mutations derived from horseshoe bat ACE2 orthologs enhance ACE2-Fc neutralization of SARS-CoV-2.PLoS Pathog. 2021 Apr 9;17(4):e1009501. doi: 10.1371/journal.ppat.1009501. eCollection 2021 Apr. PLoS Pathog. 2021. PMID: 33836016 Free PMC article.
-
Coronavirus Disease 2019: Coronaviruses and Kidney Injury.J Urol. 2020 Nov;204(5):918-925. doi: 10.1097/JU.0000000000001289. Epub 2020 Jul 17. J Urol. 2020. PMID: 32693711 Review.
-
[Source of the COVID-19 pandemic: ecology and genetics of coronaviruses (Betacoronavirus: Coronaviridae) SARS-CoV, SARS-CoV-2 (subgenus Sarbecovirus), and MERS-CoV (subgenus Merbecovirus).].Vopr Virusol. 2020;65(2):62-70. doi: 10.36233/0507-4088-2020-65-2-62-70. Vopr Virusol. 2020. PMID: 32515561 Review. Russian.
Cited by
-
The Role of Extracellular Vesicles in SARS-CoV-2-Induced Acute Kidney Injury: An Overview.Life (Basel). 2024 Jan 23;14(2):163. doi: 10.3390/life14020163. Life (Basel). 2024. PMID: 38398672 Free PMC article. Review.
-
miR-142 Targets TIM-1 in Human Endothelial Cells: Potential Implications for Stroke, COVID-19, Zika, Ebola, Dengue, and Other Viral Infections.Int J Mol Sci. 2022 Sep 6;23(18):10242. doi: 10.3390/ijms231810242. Int J Mol Sci. 2022. PMID: 36142146 Free PMC article.
-
Immune response and potential therapeutic strategies for the SARS-CoV-2 associated with the COVID-19 pandemic.Int J Biol Sci. 2022 Feb 14;18(5):1865-1877. doi: 10.7150/ijbs.66369. eCollection 2022. Int J Biol Sci. 2022. PMID: 35342348 Free PMC article. Review.
-
Catechins: Therapeutic Perspectives in COVID-19-Associated Acute Kidney Injury.Molecules. 2021 Sep 30;26(19):5951. doi: 10.3390/molecules26195951. Molecules. 2021. PMID: 34641495 Free PMC article. Review.
-
A renal YY1-KIM1-DR5 axis regulates the progression of acute kidney injury.Nat Commun. 2023 Jul 17;14(1):4261. doi: 10.1038/s41467-023-40036-z. Nat Commun. 2023. PMID: 37460623 Free PMC article.
References
-
- Chen H., Wan D.Y., Wang L., et al. (2015). Apelin protects against acute renal injury by inhibiting TGF-β1. Biochim. Biophys. Acta 1852, 1278–1287. - PubMed
-
- Chen Y., Yang D., Cheng B., et al. (2020). Clinical characteristics and outcomes of patients with diabetes and COVID-19 in association with glucose-lowering medication. Diabetes Care 43, 1399–1407. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

