A Novel Iflavirus Was Discovered in Green Rice Leafhopper Nephotettix cincticeps and Its Proliferation Was Inhibited by Infection of Rice Dwarf Virus
- PMID: 33488564
- PMCID: PMC7820178
- DOI: 10.3389/fmicb.2020.621141
A Novel Iflavirus Was Discovered in Green Rice Leafhopper Nephotettix cincticeps and Its Proliferation Was Inhibited by Infection of Rice Dwarf Virus
Abstract
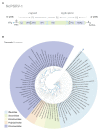
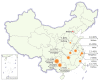
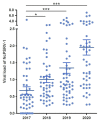
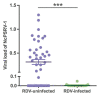
The green rice leafhopper, Nephotettix cincticeps (Hemiptera: Cicadellidae), is a key insect vector transmitting rice dwarf virus (RDV) that causes rice dwarf disease. We discovered a novel iflavirus from the transcriptomes of N. cincticeps and named it as Nephotettix cincticeps positive-stranded RNA virus-1 (NcPSRV-1). The viral genome consists of 10,524 nucleotides excluding the poly(A) tail and contains one predicted open reading frame encoding a polyprotein of 3,192 amino acids, flanked by 5' and 3' untranslated regions. NcPSRV-1 has a typical iflavirus genome arrangement and is clustered with the members of the family Iflaviridae in the phylogenetic analysis. NcPSRV-1 was detected in all tested tissues and life stages of N. cincticeps and could be transmitted horizontally and vertically. Moreover, NcPSRV-1 had high prevalence in the laboratory populations and was widely spread in field populations of N. cincticeps. NcPSRV-1 could also infect the two-striped leafhopper, Nephotettix apicalis, at a 3.33% infection rate, but was absent in the zigzag leafhopper, Recilia dorsalis, and rice Oryza sativa variety TN1. The infection of RDV altered the viral load and infection rate of NcPSRV-1 in N. cincticeps, for which it seems that RDV has an antagonistic effect on NcPSRV-1 infection in the host.
Keywords: Iflaviridae; Nephotettix cincticeps; co-infection; covert infection; distribution; rice dwarf virus; transmission; virus-virus interaction.
Copyright © 2021 Jia, Wang, Li, Chang, Yang, Yao, Bao, Song and Ye.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

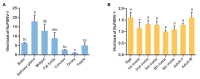


Similar articles
-
Identification, characterization and prevalence in southern China of a new iflavirus in the leafhopper Recilia dorsalis (Hemiptera: Cicadellidae).Virus Res. 2023 Jan 2;323:199005. doi: 10.1016/j.virusres.2022.199005. Epub 2022 Nov 21. Virus Res. 2023. PMID: 36410611 Free PMC article.
-
Identification and characterization of a novel rhabdovirus in green rice leafhopper, Nephotettix cincticeps.Virus Res. 2021 Apr 15;296:198281. doi: 10.1016/j.virusres.2020.198281. Epub 2021 Feb 3. Virus Res. 2021. PMID: 33548414
-
RNA interference-mediated knockdown and virus-induced suppression of Troponin C gene adversely affect the behavior or fitness of the green rice leafhopper, Nephotettix cincticeps.Arch Insect Biochem Physiol. 2018 Feb;97(2). doi: 10.1002/arch.21438. Epub 2017 Nov 30. Arch Insect Biochem Physiol. 2018. PMID: 29193300
-
Interaction between non-structural protein Pns10 of rice dwarf virus and cytoplasmic actin of leafhoppers is correlated with insect vector specificity.J Gen Virol. 2015 Apr;96(Pt 4):933-938. doi: 10.1099/jgv.0.000022. Epub 2014 Dec 12. J Gen Virol. 2015. PMID: 25502650
-
Rice Stripe Mosaic Disease: Characteristics and Control Strategies.Front Microbiol. 2021 Jul 29;12:715223. doi: 10.3389/fmicb.2021.715223. eCollection 2021. Front Microbiol. 2021. PMID: 34394065 Free PMC article. Review.
Cited by
-
Two Novel Iflaviruses Discovered in Bat Samples in Washington State.Viruses. 2022 May 7;14(5):994. doi: 10.3390/v14050994. Viruses. 2022. PMID: 35632735 Free PMC article.
-
Diversity of RNA viruses in agricultural insects.Comput Struct Biotechnol J. 2023 Sep 3;21:4312-4321. doi: 10.1016/j.csbj.2023.08.036. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37711182 Free PMC article. Review.
-
Identification and Characterization of Four Novel Viruses in Balclutha incisa.Insects. 2024 Oct 6;15(10):772. doi: 10.3390/insects15100772. Insects. 2024. PMID: 39452348 Free PMC article.
-
Identification and Characterization of Three Novel Solemo-like Viruses in the White-Backed Planthopper, Sogatella furcifera.Insects. 2024 May 28;15(6):394. doi: 10.3390/insects15060394. Insects. 2024. PMID: 38921109 Free PMC article.
-
Virus Diversity, Abundance, and Evolution in Three Different Bat Colonies in Switzerland.Viruses. 2022 Aug 29;14(9):1911. doi: 10.3390/v14091911. Viruses. 2022. PMID: 36146717 Free PMC article.
References
-
- Carreck N., Ball B., Martin S. (2010). Honey bee colony collapse and changes in viral prevalence associated with Varroa destructor. J. Apic. Res. 49, 93–94. 10.3896/IBRA.1.49.1.13 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

