Phase separation and DAXX redistribution contribute to LANA nuclear body and KSHV genome dynamics during latency and reactivation
- PMID: 33471863
- PMCID: PMC7943007
- DOI: 10.1371/journal.ppat.1009231
Phase separation and DAXX redistribution contribute to LANA nuclear body and KSHV genome dynamics during latency and reactivation
Abstract
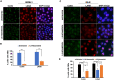
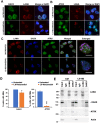
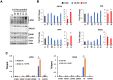
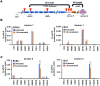
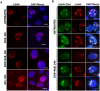
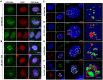
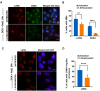
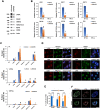
Liquid-liquid phase separation (LLPS) can drive formation of diverse and essential macromolecular structures, including those specified by viruses. Kaposi's Sarcoma-Associated Herpesvirus (KSHV) genomes associate with the viral encoded Latency-Associated Nuclear Antigen (LANA) to form stable nuclear bodies (NBs) during latent infection. Here, we show that LANA-NB formation and KSHV genome conformation involves LLPS. Using LLPS disrupting solvents, we show that LANA-NBs are partially disrupted, while DAXX and PML foci are highly resistant. LLPS disruption altered the LANA-dependent KSHV chromosome conformation but did not stimulate lytic reactivation. We found that LANA-NBs undergo major morphological transformation during KSHV lytic reactivation to form LANA-associated replication compartments encompassing KSHV DNA. DAXX colocalizes with the LANA-NBs during latency but is evicted from the LANA-associated lytic replication compartments. These findings indicate the LANA-NBs are dynamic super-molecular nuclear structures that partly depend on LLPS and undergo morphological transitions corresponding to the different modes of viral replication.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: PML is a founder and consultant for Vironika, LLC. All other authors have no Competing Interests declarations.
Figures
Similar articles
-
Site-specific association with host and viral chromatin by Kaposi's sarcoma-associated herpesvirus LANA and its reversal during lytic reactivation.J Virol. 2014 Jun;88(12):6762-77. doi: 10.1128/JVI.00268-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696474 Free PMC article.
-
Full-Length Isoforms of Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen Accumulate in the Cytoplasm of Cells Undergoing the Lytic Cycle of Replication.J Virol. 2017 Nov 30;91(24):e01532-17. doi: 10.1128/JVI.01532-17. Print 2017 Dec 15. J Virol. 2017. PMID: 28978712 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus LANA-Adjacent Regions with Distinct Functions in Episome Segregation or Maintenance.J Virol. 2019 Mar 5;93(6):e02158-18. doi: 10.1128/JVI.02158-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626680 Free PMC article.
-
KSHV Genome Replication and Maintenance in Latency.Adv Exp Med Biol. 2018;1045:299-320. doi: 10.1007/978-981-10-7230-7_14. Adv Exp Med Biol. 2018. PMID: 29896673 Review.
-
Regulation of KSHV Latency and Lytic Reactivation.Viruses. 2020 Sep 17;12(9):1034. doi: 10.3390/v12091034. Viruses. 2020. PMID: 32957532 Free PMC article. Review.
Cited by
-
Phase separations in oncogenesis, tumor progressions and metastasis: a glance from hallmarks of cancer.J Hematol Oncol. 2023 Dec 18;16(1):123. doi: 10.1186/s13045-023-01522-5. J Hematol Oncol. 2023. PMID: 38110976 Free PMC article. Review.
-
Biomolecular phase separation in stress granule assembly and virus infection.Acta Biochim Biophys Sin (Shanghai). 2023 Jul 3;55(7):1099-1118. doi: 10.3724/abbs.2023117. Acta Biochim Biophys Sin (Shanghai). 2023. PMID: 37401177 Free PMC article.
-
Molecular mechanisms and cellular functions of liquid-liquid phase separation during antiviral immune responses.Front Immunol. 2023 May 10;14:1162211. doi: 10.3389/fimmu.2023.1162211. eCollection 2023. Front Immunol. 2023. PMID: 37251408 Free PMC article. Review.
-
Viral dew: Phase separation and the formation of viral replication compartments.PLoS Pathog. 2023 Feb 16;19(2):e1011145. doi: 10.1371/journal.ppat.1011145. eCollection 2023 Feb. PLoS Pathog. 2023. PMID: 36795674 Free PMC article. No abstract available.
-
Phase separation in cancer at a glance.J Transl Med. 2023 Apr 1;21(1):237. doi: 10.1186/s12967-023-04082-x. J Transl Med. 2023. PMID: 37005672 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

