Plasma membrane lipid scrambling causing phosphatidylserine exposure negatively regulates NK cell activation
- PMID: 33469162
- PMCID: PMC8027605
- DOI: 10.1038/s41423-020-00600-9
Plasma membrane lipid scrambling causing phosphatidylserine exposure negatively regulates NK cell activation
Abstract
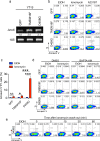
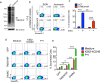
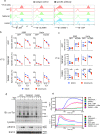
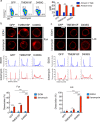
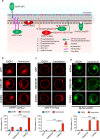
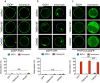
One of the hallmarks of live cells is the asymmetric distribution of lipids across their plasma membrane. Changes in this asymmetry due to lipid "scrambling" result in phosphatidylserine exposure at the cell surface that is detected by annexin V staining. This alteration is observed during cell death processes such as apoptosis, and during physiological responses such as platelet degranulation and membrane repair. Previous studies have shown that activation of NK cells is accompanied by exposure of phosphatidylserine at the cell surface. While this response was thought to be indicative of ongoing NK cell death, it may also reflect the regulation of NK cell activation in the absence of cell death. Herein, we found that NK cell activation was accompanied by rapid phosphatidylserine exposure to an extent proportional to the degree of NK cell activation. Through enforced expression of a lipid scramblase, we provided evidence that activation-induced lipid scrambling in NK cells is reversible and does not lead to cell death. In contrast, lipid scrambling attenuates NK cell activation. This response was accompanied by reduced cell surface expression of activating receptors such as 2B4, and by loss of binding of Src family protein tyrosine kinases Fyn and Lck to the inner leaflet of the plasma membrane. Hence, lipid scrambling during NK cell activation is, at least in part, a physiological response that reduces the NK cell activation level. This effect is due to the ability of lipid scrambling to alter the distribution of membrane-associated receptors and kinases required for NK cell activation.
Keywords: Lipid scrambling; NK cell activation; Phosphatidylserine exposure; Signaling; TMEM16F.
Conflict of interest statement
A.V. received a contract from Bristol Myers-Squibb to study the mechanism of action of the anti-SLAMF7 monoclonal antibody elotuzumab in multiple myeloma. He was also a consultant for Boehringer-Ingelheim on the topic of the SIRPα-CD47 blockade in anticancer immunotherapy. The authors declare no competing interests.
Figures

Similar articles
-
Critical Role of Lipid Scramblase TMEM16F in Phosphatidylserine Exposure and Repair of Plasma Membrane after Pore Formation.Cell Rep. 2020 Jan 28;30(4):1129-1140.e5. doi: 10.1016/j.celrep.2019.12.066. Cell Rep. 2020. PMID: 31995754 Free PMC article.
-
Regulation of TMEM16A/ANO1 and TMEM16F/ANO6 ion currents and phospholipid scrambling by Ca2+ and plasma membrane lipid.J Physiol. 2018 Jan 15;596(2):217-229. doi: 10.1113/JP275175. Epub 2017 Dec 18. J Physiol. 2018. PMID: 29134661 Free PMC article.
-
Regulation of Ly49D/DAP12 signal transduction by Src-family kinases and CD45.J Immunol. 2006 Jun 1;176(11):6615-23. doi: 10.4049/jimmunol.176.11.6615. J Immunol. 2006. PMID: 16709819
-
Phospholipid scramblase: an update.FEBS Lett. 2010 Jul 2;584(13):2724-30. doi: 10.1016/j.febslet.2010.03.020. Epub 2010 Mar 17. FEBS Lett. 2010. PMID: 20302864 Review.
-
Beyond apoptosis: the mechanism and function of phosphatidylserine asymmetry in the membrane of activating mast cells.Bioarchitecture. 2014;4(4-5):127-37. doi: 10.1080/19490992.2014.995516. Bioarchitecture. 2014. PMID: 25759911 Free PMC article. Review.
Cited by
-
TMEM16F scramblase regulates angiogenesis via endothelial intracellular signaling.J Cell Sci. 2024 Jul 15;137(14):jcs261566. doi: 10.1242/jcs.261566. Epub 2024 Jul 18. J Cell Sci. 2024. PMID: 38940198
-
Unique metabolism and protein expression signature in human decidual NK cells.Front Immunol. 2023 Mar 3;14:1136652. doi: 10.3389/fimmu.2023.1136652. eCollection 2023. Front Immunol. 2023. PMID: 36936959 Free PMC article.
-
Lipid scrambling in immunology: why it is important.Cell Mol Immunol. 2023 Sep;20(9):1081-1083. doi: 10.1038/s41423-023-01009-w. Epub 2023 Apr 4. Cell Mol Immunol. 2023. PMID: 37012395 Free PMC article. No abstract available.
-
FYN: emerging biological roles and potential therapeutic targets in cancer.J Transl Med. 2023 Feb 5;21(1):84. doi: 10.1186/s12967-023-03930-0. J Transl Med. 2023. PMID: 36740671 Free PMC article. Review.
-
TMEM16F Expressed in Kupffer Cells Regulates Liver Inflammation and Metabolism to Protect Against Listeria Monocytogenes.Adv Sci (Weinh). 2024 Oct;11(39):e2402693. doi: 10.1002/advs.202402693. Epub 2024 Aug 13. Adv Sci (Weinh). 2024. PMID: 39136057 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

