IMITATION SWITCH is required for normal chromatin structure and gene repression in PRC2 target domains
- PMID: 33468665
- PMCID: PMC7848606
- DOI: 10.1073/pnas.2010003118
IMITATION SWITCH is required for normal chromatin structure and gene repression in PRC2 target domains
Abstract
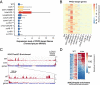
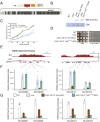
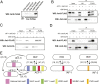
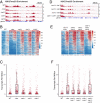
Polycomb Group (PcG) proteins are part of an epigenetic cell memory system that plays essential roles in multicellular development, stem cell biology, X chromosome inactivation, and cancer. In animals, plants, and many fungi, Polycomb Repressive Complex 2 (PRC2) catalyzes trimethylation of histone H3 lysine 27 (H3K27me3) to assemble transcriptionally repressed facultative heterochromatin. PRC2 is structurally and functionally conserved in the model fungus Neurospora crassa, and recent work in this organism has generated insights into PRC2 control and function. To identify components of the facultative heterochromatin pathway, we performed a targeted screen of Neurospora deletion strains lacking individual ATP-dependent chromatin remodeling enzymes. We found the Neurospora homolog of IMITATION SWITCH (ISW) is critical for normal transcriptional repression, nucleosome organization, and establishment of typical histone methylation patterns in facultative heterochromatin domains. We also found that stable interaction between PRC2 and chromatin depends on ISW. A functional ISW ATPase domain is required for gene repression and normal H3K27 methylation. ISW homologs interact with accessory proteins to form multiple complexes with distinct functions. Using proteomics and molecular approaches, we identified three distinct Neurospora ISW-containing complexes. A triple mutant lacking three ISW accessory factors and disrupting multiple ISW complexes led to widespread up-regulation of PRC2 target genes and altered H3K27 methylation patterns, similar to an ISW-deficient strain. Taken together, our data show that ISW is a key component of the facultative heterochromatin pathway in Neurospora, and that distinct ISW complexes perform an apparently overlapping role to regulate chromatin structure and gene repression at PRC2 target domains.
Keywords: H3K27me3; IMITATION SWITCH (ISWI); Polycomb Repressive Complex 2 (PRC2); chromatin remodeling; facultative heterochromatin.
Conflict of interest statement
The authors declare no competing interest.
Figures
Similar articles
-
The ACF chromatin-remodeling complex is essential for Polycomb repression.Elife. 2022 Mar 8;11:e77595. doi: 10.7554/eLife.77595. Elife. 2022. PMID: 35257662 Free PMC article.
-
Identification of a PRC2 Accessory Subunit Required for Subtelomeric H3K27 Methylation in Neurospora crassa.Mol Cell Biol. 2020 May 14;40(11):e00003-20. doi: 10.1128/MCB.00003-20. Print 2020 May 14. Mol Cell Biol. 2020. PMID: 32179551 Free PMC article.
-
Polycomb Repression without Bristles: Facultative Heterochromatin and Genome Stability in Fungi.Genes (Basel). 2020 Jun 9;11(6):638. doi: 10.3390/genes11060638. Genes (Basel). 2020. PMID: 32527036 Free PMC article. Review.
-
Kicking against the PRCs - A Domesticated Transposase Antagonises Silencing Mediated by Polycomb Group Proteins and Is an Accessory Component of Polycomb Repressive Complex 2.PLoS Genet. 2015 Dec 7;11(12):e1005660. doi: 10.1371/journal.pgen.1005660. eCollection 2015 Dec. PLoS Genet. 2015. PMID: 26642436 Free PMC article.
-
H3K27 methylation: a promiscuous repressive chromatin mark.Curr Opin Genet Dev. 2017 Apr;43:31-37. doi: 10.1016/j.gde.2016.11.001. Epub 2016 Dec 8. Curr Opin Genet Dev. 2017. PMID: 27940208 Free PMC article. Review.
Cited by
-
Nuclear genome organization in fungi: from gene folding to Rabl chromosomes.FEMS Microbiol Rev. 2023 May 19;47(3):fuad021. doi: 10.1093/femsre/fuad021. FEMS Microbiol Rev. 2023. PMID: 37197899 Free PMC article.
-
Linking transcriptional silencing with chromatin remodeling, folding, and positioning in the nucleus.Curr Opin Plant Biol. 2022 Oct;69:102261. doi: 10.1016/j.pbi.2022.102261. Epub 2022 Jul 13. Curr Opin Plant Biol. 2022. PMID: 35841650 Free PMC article. Review.
-
The Upstream Sequence Transcription Complex dictates nucleosome positioning and promoter accessibility at piRNA genes in the C. elegans germ line.PLoS Genet. 2024 Jul 10;20(7):e1011345. doi: 10.1371/journal.pgen.1011345. eCollection 2024 Jul. PLoS Genet. 2024. PMID: 38985845 Free PMC article.
-
Interplay between acetylation and ubiquitination of imitation switch chromatin remodeler Isw1 confers multidrug resistance in Cryptococcus neoformans.Elife. 2024 Jan 22;13:e85728. doi: 10.7554/eLife.85728. Elife. 2024. PMID: 38251723 Free PMC article.
-
A crucial role for dynamic expression of components encoding the negative arm of the circadian clock.Nat Commun. 2023 Jun 8;14(1):3371. doi: 10.1038/s41467-023-38817-7. Nat Commun. 2023. PMID: 37291101 Free PMC article.
References
-
- Schuettengruber B., Bourbon H.-M., Di Croce L., Cavalli G., Genome regulation by polycomb and trithorax: 70 years and counting. Cell 171, 34–57 (2017). - PubMed
-
- Cao R., et al. , Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043 (2002). - PubMed
-
- Czermin B., et al. , Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111, 185–196 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

