Origin, Regulation, and Fitness Effect of Chromosomal Rearrangements in the Yeast Saccharomyces cerevisiae
- PMID: 33466757
- PMCID: PMC7830279
- DOI: 10.3390/ijms22020786
Origin, Regulation, and Fitness Effect of Chromosomal Rearrangements in the Yeast Saccharomyces cerevisiae
Abstract
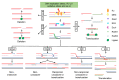
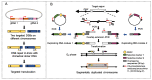
Chromosomal rearrangements comprise unbalanced structural variations resulting in gain or loss of DNA copy numbers, as well as balanced events including translocation and inversion that are copy number neutral, both of which contribute to phenotypic evolution in organisms. The exquisite genetic assay and gene editing tools available for the model organism Saccharomyces cerevisiae facilitate deep exploration of the mechanisms underlying chromosomal rearrangements. We discuss here the pathways and influential factors of chromosomal rearrangements in S. cerevisiae. Several methods have been developed to generate on-demand chromosomal rearrangements and map the breakpoints of rearrangement events. Finally, we highlight the contributions of chromosomal rearrangements to drive phenotypic evolution in various S. cerevisiae strains. Given the evolutionary conservation of DNA replication and recombination in organisms, the knowledge gathered in the small genome of yeast can be extended to the genomes of higher eukaryotes.
Keywords: DNA repair; S. cerevisiae; chromosomal rearrangement; recombination; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Mechanisms underlying genome instability mediated by formation of foldback inversions in Saccharomyces cerevisiae.Elife. 2020 Aug 7;9:e58223. doi: 10.7554/eLife.58223. Elife. 2020. PMID: 32762846 Free PMC article.
-
Genome-wide amplifications caused by chromosomal rearrangements play a major role in the adaptive evolution of natural yeast.Genetics. 2003 Dec;165(4):1745-59. doi: 10.1093/genetics/165.4.1745. Genetics. 2003. PMID: 14704163 Free PMC article.
-
Different aneuploidies arise from the same bridge-induced chromosomal translocation event in Saccharomyces cerevisiae.Genetics. 2010 Nov;186(3):775-90. doi: 10.1534/genetics.110.120683. Epub 2010 Aug 30. Genetics. 2010. PMID: 20805555 Free PMC article.
-
Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae.DNA Repair (Amst). 2006 Sep 8;5(9-10):1010-20. doi: 10.1016/j.dnarep.2006.05.027. Epub 2006 Jun 23. DNA Repair (Amst). 2006. PMID: 16798113 Review.
-
Faithful after break-up: suppression of chromosomal translocations.Cell Mol Life Sci. 2009 Oct;66(19):3149-60. doi: 10.1007/s00018-009-0068-5. Epub 2009 Jun 23. Cell Mol Life Sci. 2009. PMID: 19547915 Free PMC article. Review.
Cited by
-
Large-scale genomic rearrangements boost SCRaMbLE in Saccharomyces cerevisiae.Nat Commun. 2024 Jan 26;15(1):770. doi: 10.1038/s41467-023-44511-5. Nat Commun. 2024. PMID: 38278805 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

