Deciphering the LRRK code: LRRK1 and LRRK2 phosphorylate distinct Rab proteins and are regulated by diverse mechanisms
- PMID: 33459343
- PMCID: PMC7886321
- DOI: 10.1042/BCJ20200937
Deciphering the LRRK code: LRRK1 and LRRK2 phosphorylate distinct Rab proteins and are regulated by diverse mechanisms
Abstract
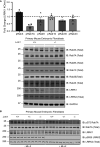
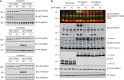
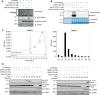
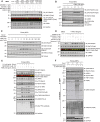
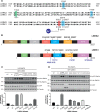
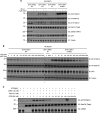
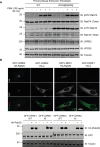
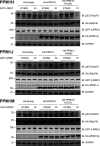
Autosomal dominant mutations in LRRK2 that enhance kinase activity cause Parkinson's disease. LRRK2 phosphorylates a subset of Rab GTPases including Rab8A and Rab10 within its effector binding motif. Here, we explore whether LRRK1, a less studied homolog of LRRK2 that regulates growth factor receptor trafficking and osteoclast biology might also phosphorylate Rab proteins. Using mass spectrometry, we found that in LRRK1 knock-out cells, phosphorylation of Rab7A at Ser72 was most impacted. This residue lies at the equivalent site targeted by LRRK2 on Rab8A and Rab10. Accordingly, recombinant LRRK1 efficiently phosphorylated Rab7A at Ser72, but not Rab8A or Rab10. Employing a novel phospho-specific antibody, we found that phorbol ester stimulation of mouse embryonic fibroblasts markedly enhanced phosphorylation of Rab7A at Ser72 via LRRK1. We identify two LRRK1 mutations (K746G and I1412T), equivalent to the LRRK2 R1441G and I2020T Parkinson's mutations, that enhance LRRK1 mediated phosphorylation of Rab7A. We demonstrate that two regulators of LRRK2 namely Rab29 and VPS35[D620N], do not influence LRRK1. Widely used LRRK2 inhibitors do not inhibit LRRK1, but we identify a promiscuous inhibitor termed GZD-824 that inhibits both LRRK1 and LRRK2. The PPM1H Rab phosphatase when overexpressed dephosphorylates Rab7A. Finally, the interaction of Rab7A with its effector RILP is not affected by LRRK1 phosphorylation and we observe that maximal stimulation of the TBK1 or PINK1 pathway does not elevate Rab7A phosphorylation. Altogether, these findings reinforce the idea that the LRRK enzymes have evolved as major regulators of Rab biology with distinct substrate specificity.
Keywords: Rab GTPase; kinase; leucine rich repeat kinase; phosphorylation.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures

Similar articles
-
Development of phospho-specific Rab protein antibodies to monitor in vivo activity of the LRRK2 Parkinson's disease kinase.Biochem J. 2018 Jan 2;475(1):1-22. doi: 10.1042/BCJ20170802. Biochem J. 2018. PMID: 29127256 Free PMC article.
-
The Parkinson's disease VPS35[D620N] mutation enhances LRRK2-mediated Rab protein phosphorylation in mouse and human.Biochem J. 2018 Jun 6;475(11):1861-1883. doi: 10.1042/BCJ20180248. Biochem J. 2018. PMID: 29743203 Free PMC article.
-
PPM1H phosphatase counteracts LRRK2 signaling by selectively dephosphorylating Rab proteins.Elife. 2019 Oct 30;8:e50416. doi: 10.7554/eLife.50416. Elife. 2019. PMID: 31663853 Free PMC article.
-
LRRK2 phosphorylation of Rab GTPases in Parkinson's disease.FEBS Lett. 2023 Mar;597(6):811-818. doi: 10.1002/1873-3468.14492. Epub 2022 Sep 26. FEBS Lett. 2023. PMID: 36114007 Review.
-
LRRK2 and Rab GTPases.Biochem Soc Trans. 2018 Dec 17;46(6):1707-1712. doi: 10.1042/BST20180470. Epub 2018 Nov 22. Biochem Soc Trans. 2018. PMID: 30467121 Review.
Cited by
-
Genome-wide screen reveals Rab12 GTPase as a critical activator of Parkinson's disease-linked LRRK2 kinase.Elife. 2023 Oct 24;12:e87098. doi: 10.7554/eLife.87098. Elife. 2023. PMID: 37874635 Free PMC article.
-
The Regulation of Rab GTPases by Phosphorylation.Biomolecules. 2021 Sep 10;11(9):1340. doi: 10.3390/biom11091340. Biomolecules. 2021. PMID: 34572553 Free PMC article. Review.
-
Increased LRRK2 kinase activity alters neuronal autophagy by disrupting the axonal transport of autophagosomes.Curr Biol. 2021 May 24;31(10):2140-2154.e6. doi: 10.1016/j.cub.2021.02.061. Epub 2021 Mar 24. Curr Biol. 2021. PMID: 33765413 Free PMC article.
-
Protein kinase C showcases allosteric control: activation of LRRK1.Biochem J. 2023 Feb 14;480(3):219-223. doi: 10.1042/BCJ20220507. Biochem J. 2023. PMID: 36762701 Free PMC article.
-
LRRK2 and idiopathic Parkinson's disease.Trends Neurosci. 2022 Mar;45(3):224-236. doi: 10.1016/j.tins.2021.12.002. Epub 2022 Jan 4. Trends Neurosci. 2022. PMID: 34991886 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

