Structural variability and differentiation of niches in the rhizosphere and endosphere bacterial microbiome of moso bamboo (Phyllostachys edulis)
- PMID: 33452327
- PMCID: PMC7810855
- DOI: 10.1038/s41598-021-80971-9
Structural variability and differentiation of niches in the rhizosphere and endosphere bacterial microbiome of moso bamboo (Phyllostachys edulis)
Abstract
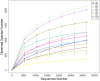
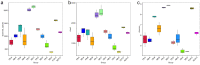
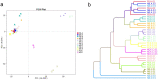
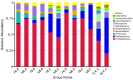
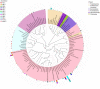
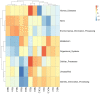
The plant microbiota play a key role in plant productivity, nutrient uptake, resistance to stress and flowering. The flowering of moso bamboo has been a focus of study. The mechanism of flowering is related to nutrient uptake, temperature, hormone balance and regulation of key genes. However, the connection between microbiota of moso bamboo and its flowering is unknown. In this study, samples of rhizosphere soil, rhizomes, roots and leaves of flowering and nonflowering plants were collected, and 16S rRNA amplicon Illumina sequencing was utilized to separate the bacterial communities associated with different flowering stages of moso bamboo. We identified 5442 OTUs, and the number of rhizosphere soil OTUs was much higher than those of other samples. Principal component analysis (PCA) and hierarchical clustering (Bray Curtis dis) analysis revealed that the bacterial microorganisms related to rhizosphere soil and endophytic tissues of moso bamboo differed significantly from those in bulk soil and rhizobacterial and endosphere microbiomes. In addition, the PCA analyses of root and rhizosphere soil revealed different structures of microbial communities between bamboo that is flowering and not flowering. Through the analysis of core microorganisms, it was found that Flavobacterium, Bacillus and Stenotrophomonas played an important role in the absorption of N elements, which may affect the flowering time of moso bamboo. Our results delineate the complex host-microbe interactions of this plant. We also discuss the potential influence of bacterial microbiome in flowering, which can provide a basis for the development and utilization of moso bamboo.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Assembly patterns and key taxa of bacterial communities in the rhizosphere soil of moso bamboo (Phyllostachys pubescens) under different Cd and Pb pollution.Int J Phytoremediation. 2024 Sep;26(11):1776-1786. doi: 10.1080/15226514.2024.2356204. Epub 2024 May 23. Int J Phytoremediation. 2024. PMID: 38780520
-
Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees.Microbiome. 2017 Feb 23;5(1):25. doi: 10.1186/s40168-017-0241-2. Microbiome. 2017. PMID: 28231859 Free PMC article.
-
Dynamics of rhizosphere microbial structure and function associated with the biennial bearing of moso bamboo.J Environ Manage. 2024 Feb;351:119977. doi: 10.1016/j.jenvman.2023.119977. Epub 2023 Dec 30. J Environ Manage. 2024. PMID: 38160549
-
Engineering agricultural soil microbiomes and predicting plant phenotypes.Trends Microbiol. 2024 Sep;32(9):858-873. doi: 10.1016/j.tim.2024.02.003. Epub 2024 Feb 29. Trends Microbiol. 2024. PMID: 38429182 Review.
-
Transcriptomic Complexity of Culm Growth and Development in Different Types of Moso Bamboo.Int J Mol Sci. 2023 Apr 18;24(8):7425. doi: 10.3390/ijms24087425. Int J Mol Sci. 2023. PMID: 37108588 Free PMC article. Review.
Cited by
-
Temporally Selective Modification of the Tomato Rhizosphere and Root Microbiome by Volcanic Ash Fertilizer Containing Micronutrients.Appl Environ Microbiol. 2022 Apr 12;88(7):e0004922. doi: 10.1128/aem.00049-22. Epub 2022 Mar 21. Appl Environ Microbiol. 2022. PMID: 35311513 Free PMC article.
-
Nodule-associated diazotrophic community succession is driven by developmental phases combined with microhabitat of Sophora davidii.Front Microbiol. 2022 Dec 1;13:1078208. doi: 10.3389/fmicb.2022.1078208. eCollection 2022. Front Microbiol. 2022. PMID: 36532429 Free PMC article.
-
Bacterial community and culturable actinomycetes of Phyllostachys viridiglaucescens rhizosphere.Antonie Van Leeuwenhoek. 2024 Jan 3;117(1):9. doi: 10.1007/s10482-023-01906-0. Antonie Van Leeuwenhoek. 2024. PMID: 38170239
-
Seasonal patterns of rhizosphere microorganisms suggest carbohydrate-degrading and nitrogen-fixing microbes contribute to the attribute of full-year shooting in woody bamboo Cephalostachyum pingbianense.Front Microbiol. 2022 Nov 29;13:1033293. doi: 10.3389/fmicb.2022.1033293. eCollection 2022. Front Microbiol. 2022. PMID: 36523824 Free PMC article.
-
Bacterial community structure of the sunflower (Helianthus annuus) endosphere.Plant Signal Behav. 2021 Dec 2;16(12):1974217. doi: 10.1080/15592324.2021.1974217. Epub 2021 Sep 30. Plant Signal Behav. 2021. PMID: 34590546 Free PMC article.
References
-
- Chaparro JM, Sheflin AM, Manter DK, Vivanco JM. Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils. 2012;48:489–499. doi: 10.1007/s00374-012-0691-4. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

