Involvement of SUT1 and SUT2 Sugar Transporters in the Impairment of Sugar Transport and Changes in Phloem Exudate Contents in Phytoplasma-Infected Plants
- PMID: 33451049
- PMCID: PMC7828548
- DOI: 10.3390/ijms22020745
Involvement of SUT1 and SUT2 Sugar Transporters in the Impairment of Sugar Transport and Changes in Phloem Exudate Contents in Phytoplasma-Infected Plants
Abstract
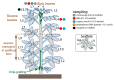
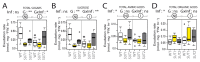
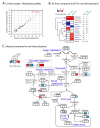
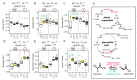
Phytoplasmas inhabit phloem sieve elements and cause abnormal growth and altered sugar partitioning. However, how they interact with phloem functions is not clearly known. The phloem responses were investigated in tomatoes infected by "Candidatus Phytoplasma solani" at the beginning of the symptomatic stage, the first symptoms appearing in the newly emerged leaf at the stem apex. Antisense lines impaired in the phloem sucrose transporters SUT1 and SUT2 were included. In symptomatic sink leaves, leaf curling was associated with higher starch accumulation and the expression of defense genes. The analysis of leaf midribs of symptomatic leaves indicated that transcript levels for genes acting in the glycolysis and peroxisome metabolism differed from these in noninfected plants. The phytoplasma also multiplied in the three lower source leaves, even if it was not associated with the symptoms. In these leaves, the rate of phloem sucrose exudation was lower for infected plants. Metabolite profiling of phloem sap-enriched exudates revealed that glycolate and aspartate levels were affected by the infection. Their levels were also affected in the noninfected SUT1- and SUT2-antisense lines. The findings suggest the role of sugar transporters in the responses to infection and describe the consequences of impaired sugar transport on the primary metabolism.
Keywords: carbon allocation; defense; glycolate; glyoxylate; metabolome; peroxisome; phloem; photorespiration; phytoplasma; plant-pathogen interaction; source-sink relationships; sugar metabolism.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
'Candidatus Phytoplasma solani' interferes with the distribution and uptake of iron in tomato.BMC Genomics. 2019 Sep 10;20(1):703. doi: 10.1186/s12864-019-6062-x. BMC Genomics. 2019. PMID: 31500568 Free PMC article.
-
Laser microdissection of grapevine leaf phloem infected by stolbur reveals site-specific gene responses associated to sucrose transport and metabolism.Plant Cell Environ. 2013 Feb;36(2):343-55. doi: 10.1111/j.1365-3040.2012.02577.x. Epub 2012 Aug 7. Plant Cell Environ. 2013. PMID: 22788215
-
Integrated Phloem Sap mRNA and Protein Expression Analysis Reveals Phytoplasma-infection Responses in Mulberry.Mol Cell Proteomics. 2018 Sep;17(9):1702-1719. doi: 10.1074/mcp.RA118.000670. Epub 2018 May 30. Mol Cell Proteomics. 2018. PMID: 29848783 Free PMC article.
-
Sugar Transporters in Plants: New Insights and Discoveries.Plant Cell Physiol. 2017 Sep 1;58(9):1442-1460. doi: 10.1093/pcp/pcx090. Plant Cell Physiol. 2017. PMID: 28922744 Review.
-
Source-To-Sink Transport of Sugar and Its Role in Male Reproductive Development.Genes (Basel). 2022 Jul 25;13(8):1323. doi: 10.3390/genes13081323. Genes (Basel). 2022. PMID: 35893060 Free PMC article. Review.
Cited by
-
Increased susceptibility to Chrysanthemum Yellows phytoplasma infection in Atcals7ko plants is accompanied by enhanced expression of carbohydrate transporters.Planta. 2022 Jul 17;256(2):43. doi: 10.1007/s00425-022-03954-8. Planta. 2022. PMID: 35842878 Free PMC article.
-
Cytokinin Regulation of Source-Sink Relationships in Plant-Pathogen Interactions.Front Plant Sci. 2021 Aug 24;12:677585. doi: 10.3389/fpls.2021.677585. eCollection 2021. Front Plant Sci. 2021. PMID: 34504504 Free PMC article. Review.
-
Spatiotemporal and Quantitative Monitoring of the Fate of "Candidatus Phytoplasma Solani" in Tomato Plants Infected by Grafting.Pathogens. 2021 Jun 26;10(7):811. doi: 10.3390/pathogens10070811. Pathogens. 2021. PMID: 34206841 Free PMC article.
-
Transcriptomic Profiling of Sugarcane White Leaf (SCWL) Canes during Maturation Phase.Plants (Basel). 2024 Jun 4;13(11):1551. doi: 10.3390/plants13111551. Plants (Basel). 2024. PMID: 38891358 Free PMC article.
-
The Interplay between Enucleated Sieve Elements and Companion Cells.Plants (Basel). 2023 Aug 23;12(17):3033. doi: 10.3390/plants12173033. Plants (Basel). 2023. PMID: 37687278 Free PMC article. Review.
References
-
- Bertaccini A., Duduk B., Paltrinieri S., Contaldo N. Phytoplasmas and phytoplasma diseases: A severe threat to agriculture. Am. J. Plant Sci. 2014;5:1763–1788. doi: 10.4236/ajps.2014.512191. - DOI
-
- Wei W., Kakizawa S., Suzuki S., Jung H.-Y., Nishigawa H., Miyata S.-I., Oshima K., Ugaki M., Hibi T., Namba S. In Planta dynamic analysis of onion yellows phytoplasma using localized inoculation by insect transmission. Phytopathology. 2004;94:244–250. doi: 10.1094/PHYTO.2004.94.3.244. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

