Molecular pathogenesis of rhegmatogenous retinal detachment
- PMID: 33441730
- PMCID: PMC7806834
- DOI: 10.1038/s41598-020-80005-w
Molecular pathogenesis of rhegmatogenous retinal detachment
Abstract
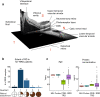
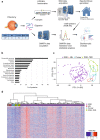
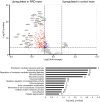
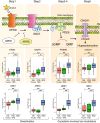
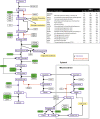
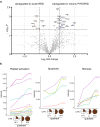
Rhegmatogenous retinal detachment (RRD) is an ophthalmic emergency, which usually requires prompt surgery to prevent further detachment and restore sensory function. Although several individual factors have been suggested, a systems level understanding of molecular pathomechanisms underlying this severe eye disorder is lacking. To address this gap in knowledge we performed the molecular level systems pathology analysis of the vitreous from 127 patients with RRD using state-of-the art quantitative mass spectrometry to identify the individual key proteins, as well as the biochemical pathways contributing to the development of the disease. RRD patients have specific vitreous proteome profiles compared to other diseases such as macular hole, pucker, or proliferative diabetic retinopathy eyes. Our data indicate that various mechanisms, including glycolysis, photoreceptor death, and Wnt and MAPK signaling, are activated during or after the RRD to promote retinal cell survival. In addition, platelet-mediated wound healing processes, cell adhesion molecules reorganization and apoptotic processes were detected during RRD progression or proliferative vitreoretinopathy formation. These findings improve the understanding of RRD pathogenesis, identify novel targets for treatment of this ophthalmic disease, and possibly affect the prognosis of eyes treated or operated upon due to RRD.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Elevated erythropoietin in vitreous of patients with rhegmatogenous retinal detachment and proliferative vitreoretinopathy.Ophthalmic Res. 2009;42(3):138-40. doi: 10.1159/000228592. Epub 2009 Jul 15. Ophthalmic Res. 2009. PMID: 19602911
-
Soluble TNF receptors in vitreoretinal proliferative disease.Invest Ophthalmol Vis Sci. 2001 Jun;42(7):1586-91. Invest Ophthalmol Vis Sci. 2001. PMID: 11381065
-
Proteomic insight into the pathogenesis of CAPN5-vitreoretinopathy.Sci Rep. 2019 May 20;9(1):7608. doi: 10.1038/s41598-019-44031-7. Sci Rep. 2019. PMID: 31110225 Free PMC article.
-
[The current data on the proliferative process in diabetic retinopathy].Vestn Oftalmol. 1999 May-Jun;115(3):37-40. Vestn Oftalmol. 1999. PMID: 10432854 Review. Russian. No abstract available.
-
Misdirected aqueous flow in rhegmatogenous retinal detachment: a pathophysiology update.Surv Ophthalmol. 2015 Jan-Feb;60(1):51-9. doi: 10.1016/j.survophthal.2014.07.002. Epub 2014 Aug 10. Surv Ophthalmol. 2015. PMID: 25223495 Review.
Cited by
-
Anti-inflammatory potential of simvastatin and amfenac in ARPE-19 cells; insights in preventing re-detachment and proliferative vitreoretinopathy after rhegmatogenous retinal detachment surgery.Int Ophthalmol. 2024 Mar 26;44(1):158. doi: 10.1007/s10792-024-03067-z. Int Ophthalmol. 2024. PMID: 38530532 Free PMC article.
-
Peripheral monocytes and neutrophils promote photoreceptor cell death in an experimental retinal detachment model.Cell Death Dis. 2023 Dec 16;14(12):834. doi: 10.1038/s41419-023-06350-6. Cell Death Dis. 2023. PMID: 38102109 Free PMC article.
-
4D label-free proteomic analysis of vitreous from patients with rhegmatogenous retinal detachment.Int J Ophthalmol. 2023 Apr 18;16(4):523-531. doi: 10.18240/ijo.2023.04.05. eCollection 2023. Int J Ophthalmol. 2023. PMID: 37077476 Free PMC article.
-
Proteomics profiling of vitreous humor reveals complement and coagulation components, adhesion factors, and neurodegeneration markers as discriminatory biomarkers of vitreoretinal eye diseases.Front Immunol. 2023 Feb 16;14:1107295. doi: 10.3389/fimmu.2023.1107295. eCollection 2023. Front Immunol. 2023. PMID: 36875133 Free PMC article.
-
Vitreous humor proteome: unraveling the molecular mechanisms underlying proliferative and neovascular vitreoretinal diseases.Cell Mol Life Sci. 2022 Dec 31;80(1):22. doi: 10.1007/s00018-022-04670-y. Cell Mol Life Sci. 2022. PMID: 36585968 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

