Reticulon-3 Promotes Endosome Maturation at ER Membrane Contact Sites
- PMID: 33434526
- PMCID: PMC7837408
- DOI: 10.1016/j.devcel.2020.12.014
Reticulon-3 Promotes Endosome Maturation at ER Membrane Contact Sites
Abstract
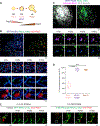
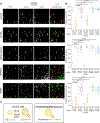
ER tubules form and maintain membrane contact sites (MCSs) with endosomes. How and why these ER-endosome MCSs persist as endosomes traffic and mature is poorly understood. Here we find that a member of the reticulon protein family, Reticulon-3L (Rtn3L), enriches at ER-endosome MCSs as endosomes mature. We show that this localization is due to the long divergent N-terminal cytoplasmic domain of Rtn3L. We found that Rtn3L is recruited to ER-endosome MCSs by endosomal protein Rab9a, which marks a transition stage between early and late endosomes. Rab9a utilizes an FSV region to recruit Rtn3L via its six LC3-interacting region motifs. Consistent with our localization results, depletion or deletion of RTN3 from cells results in endosome maturation and cargo sorting defects, similar to RAB9A depletion. Together our data identify a tubular ER protein that promotes endosome maturation at ER MCSs.
Keywords: endoplasmic reticulum; endosome maturation; membrane contact sites.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures




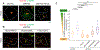
Similar articles
-
Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature.Mol Biol Cell. 2013 Apr;24(7):1030-40. doi: 10.1091/mbc.E12-10-0733. Epub 2013 Feb 6. Mol Biol Cell. 2013. PMID: 23389631 Free PMC article.
-
PDZD8 interacts with Protrudin and Rab7 at ER-late endosome membrane contact sites associated with mitochondria.Nat Commun. 2020 Jul 20;11(1):3645. doi: 10.1038/s41467-020-17451-7. Nat Commun. 2020. PMID: 32686675 Free PMC article.
-
A Novel Class of ER Membrane Proteins Regulates ER-Associated Endosome Fission.Cell. 2018 Sep 20;175(1):254-265.e14. doi: 10.1016/j.cell.2018.08.030. Epub 2018 Sep 13. Cell. 2018. PMID: 30220460 Free PMC article.
-
Touché! STARD3 and STARD3NL tether the ER to endosomes.Biochem Soc Trans. 2016 Apr 15;44(2):493-8. doi: 10.1042/BST20150269. Biochem Soc Trans. 2016. PMID: 27068960 Review.
-
Endosome biogenesis is controlled by ER and the cytoskeleton at tripartite junctions.Curr Opin Cell Biol. 2023 Feb;80:102155. doi: 10.1016/j.ceb.2023.102155. Epub 2023 Feb 26. Curr Opin Cell Biol. 2023. PMID: 36848759 Review.
Cited by
-
Multiple roles for actin in secretory and endocytic pathways.Curr Biol. 2021 May 24;31(10):R603-R618. doi: 10.1016/j.cub.2021.03.038. Curr Biol. 2021. PMID: 34033793 Free PMC article. Review.
-
ER remodeling via ER-phagy.Mol Cell. 2022 Apr 21;82(8):1492-1500. doi: 10.1016/j.molcel.2022.02.018. Mol Cell. 2022. PMID: 35452617 Free PMC article. Review.
-
Exosomes target HBV-host interactions to remodel the hepatic immune microenvironment.J Nanobiotechnology. 2024 Jun 5;22(1):315. doi: 10.1186/s12951-024-02544-y. J Nanobiotechnology. 2024. PMID: 38840207 Free PMC article. Review.
-
Membrane Curvature: The Inseparable Companion of Autophagy.Cells. 2023 Apr 11;12(8):1132. doi: 10.3390/cells12081132. Cells. 2023. PMID: 37190041 Free PMC article. Review.
-
Enhancing drug penetration in solid tumors via nanomedicine: Evaluation models, strategies and perspectives.Bioact Mater. 2023 Oct 26;32:445-472. doi: 10.1016/j.bioactmat.2023.10.017. eCollection 2024 Feb. Bioact Mater. 2023. PMID: 37965242 Free PMC article. Review.
References
-
- Bindels DS, Haarbosch L, van Weeren L, Postma M, Wiese KE, Mastop M, Aumonier S, Gotthard G, Royant A, Hink MA, et al. (2017). mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat. Methods 14, 53–56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

