Comparative plastid genomics of four Pilea (Urticaceae) species: insight into interspecific plastid genome diversity in Pilea
- PMID: 33413130
- PMCID: PMC7792329
- DOI: 10.1186/s12870-020-02793-7
Comparative plastid genomics of four Pilea (Urticaceae) species: insight into interspecific plastid genome diversity in Pilea
Abstract
Background: Pilea is a genus of perennial herbs from the family Urticaceae, and some species are used as courtyard ornamentals or for medicinal purposes. At present, there is no information about the plastid genome of Pilea, which limits our understanding of this genus. Here, we report 4 plastid genomes of Pilea taxa (Pilea mollis, Pilea glauca 'Greizy', Pilea peperomioides and Pilea serpyllacea 'Globosa') and performed comprehensive comparative analysis.
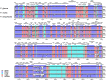
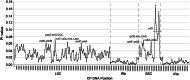
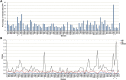
Results: The four plastid genomes all have a typical quartile structure. The lengths of the plastid genomes ranged from 150,398 bp to 152,327 bp, and each genome contained 113 unique genes, including 79 protein-coding genes, 4 rRNA genes, and 30 tRNA genes. Comparative analysis showed a rather high level of sequence divergence in the four genomes. Moreover, eight hypervariable regions were identified (petN-psbM, psbZ-trnG-GCC, trnT-UGU-trnL-UAA, accD-psbI, ndhF-rpl32, rpl32-trnL-UAG, ndhA-intron and ycf1), which are proposed for use as DNA barcode regions. Phylogenetic relationships based on the plastid genomes of 23 species of 14 genera of Urticaceae resulted in the placement of Pilea in the middle and lower part of the phylogenetic tree, with 100% bootstrap support within Urticaceae.
Conclusion: Our results enrich the resources concerning plastid genomes. Comparative plastome analysis provides insight into the interspecific diversity of the plastid genome of Pilea. The identified hypervariable regions could be used for developing molecular markers applicable in various research areas.
Keywords: Hypervariable region; Interspecific diversity; Phylogenetic relationship; Pilea; Plastid genome.
Conflict of interest statement
The authors declare that they have no competing interests
Figures




Similar articles
-
Evolutionary Analysis of Plastid Genomes of Seven Lonicera L. Species: Implications for Sequence Divergence and Phylogenetic Relationships.Int J Mol Sci. 2018 Dec 14;19(12):4039. doi: 10.3390/ijms19124039. Int J Mol Sci. 2018. PMID: 30558106 Free PMC article.
-
Plastid genomes of Elaeagnus mollis: comparative and phylogenetic analyses.J Genet. 2020;99:85. J Genet. 2020. PMID: 33361637
-
Characteristics of plastid genomes in the genus Ceratostigma inhabiting arid habitats in China and their phylogenomic implications.BMC Plant Biol. 2023 Jun 7;23(1):303. doi: 10.1186/s12870-023-04323-7. BMC Plant Biol. 2023. PMID: 37280518 Free PMC article.
-
Complete chloroplast genomes of eight Delphinium taxa (Ranunculaceae) endemic to Xinjiang, China: insights into genome structure, comparative analysis, and phylogenetic relationships.BMC Plant Biol. 2024 Jun 26;24(1):600. doi: 10.1186/s12870-024-05279-y. BMC Plant Biol. 2024. PMID: 38926811 Free PMC article.
-
Dynamic evolution of the plastome in the Elm family (Ulmaceae).Planta. 2022 Dec 17;257(1):14. doi: 10.1007/s00425-022-04045-4. Planta. 2022. PMID: 36526857 Review.
Cited by
-
Chloroplast genome of Ocimum basilicum var. purpurascens Bentham 1830 (Lamiaceae).Mitochondrial DNA B Resour. 2024 Feb 5;9(2):252-256. doi: 10.1080/23802359.2024.2310145. eCollection 2024. Mitochondrial DNA B Resour. 2024. PMID: 38328459 Free PMC article.
-
The Complete Chloroplast Genome Sequence of Laportea bulbifera (Sieb. et Zucc.) Wedd. and Comparative Analysis with Its Congeneric Species.Genes (Basel). 2022 Nov 28;13(12):2230. doi: 10.3390/genes13122230. Genes (Basel). 2022. PMID: 36553498 Free PMC article.
-
Deep Insights Into the Plastome Evolution and Phylogenetic Relationships of the Tribe Urticeae (Family Urticaceae).Front Plant Sci. 2022 May 20;13:870949. doi: 10.3389/fpls.2022.870949. eCollection 2022. Front Plant Sci. 2022. PMID: 35668809 Free PMC article.
-
The complete chloroplast genome and phylogenetic analysis of Jacobaea maritima (Asteraceae).Mitochondrial DNA B Resour. 2023 Jul 22;8(7):771-776. doi: 10.1080/23802359.2023.2238937. eCollection 2023. Mitochondrial DNA B Resour. 2023. PMID: 37492062 Free PMC article.
-
Plastid genome data provide new insights into the dynamic evolution of the tribe Ampelopsideae (Vitaceae).BMC Genomics. 2024 Mar 5;25(1):247. doi: 10.1186/s12864-024-10149-w. BMC Genomics. 2024. PMID: 38443830 Free PMC article.
References
-
- Prabhakar KR, Veerapur VP, Bansal P, Parihar VK, Reddy Kandadi M, Bhagath Kumar P, Priyadarsini KI, Unnikrishnan MK. Antioxidant and radioprotective effect of the active fraction of Pilea microphylla (L.) ethanolic extract. Chem Biol Interact. 2007;165(1):22–32. doi: 10.1016/j.cbi.2006.10.007. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

