New Insectotoxin from Tibellus Oblongus Spider Venom Presents Novel Adaptation of ICK Fold
- PMID: 33406803
- PMCID: PMC7824768
- DOI: 10.3390/toxins13010029
New Insectotoxin from Tibellus Oblongus Spider Venom Presents Novel Adaptation of ICK Fold
Abstract
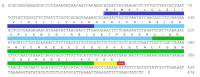
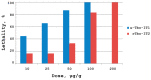
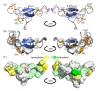
The Tibellus oblongus spider is an active predator that does not spin webs and remains poorly investigated in terms of venom composition. Here, we present a new toxin, named Tbo-IT2, predicted by cDNA analysis of venom glands transcriptome. The presence of Tbo-IT2 in the venom was confirmed by proteomic analyses using the LC-MS and MS/MS techniques. The distinctive features of Tbo-IT2 are the low similarity of primary structure with known animal toxins and the unusual motif of 10 cysteine residues distribution. Recombinant Tbo-IT2 (rTbo-IT2), produced in E. coli using the thioredoxin fusion protein strategy, was structurally and functionally studied. rTbo-IT2 showed insecticidal activity on larvae of the housefly Musca domestica (LD100 200 μg/g) and no activity on the panel of expressed neuronal receptors and ion channels. The spatial structure of the peptide was determined in a water solution by NMR spectroscopy. The Tbo-IT2 structure is a new example of evolutionary adaptation of a well-known inhibitor cystine knot (ICK) fold to 5 disulfide bonds configuration, which determines additional conformational stability and gives opportunities for insectotoxicity and probably some other interesting features.
Keywords: ICK fold; NMR structure; insectotoxin; proteome; spider venom; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





Similar articles
-
Venom-gland transcriptomics and venom proteomics of the Tibellus oblongus spider.Sci Data. 2023 Nov 22;10(1):820. doi: 10.1038/s41597-023-02703-0. Sci Data. 2023. PMID: 37993463 Free PMC article.
-
ω-Tbo-IT1-New Inhibitor of Insect Calcium Channels Isolated from Spider Venom.Sci Rep. 2015 Nov 27;5:17232. doi: 10.1038/srep17232. Sci Rep. 2015. PMID: 26611444 Free PMC article.
-
A novel ICK peptide from the Loxosceles intermedia (brown spider) venom gland: cloning, heterologous expression and immunological cross-reactivity approaches.Toxicon. 2013 Sep;71:147-58. doi: 10.1016/j.toxicon.2013.05.014. Epub 2013 Jun 8. Toxicon. 2013. PMID: 23751278
-
An overview of peptide toxins from the venom of the Chinese bird spider Selenocosmia huwena Wang [=Ornithoctonus huwena (Wang)].Toxicon. 2004 Apr;43(5):575-85. doi: 10.1016/j.toxicon.2004.02.005. Toxicon. 2004. PMID: 15066414 Review.
-
Recent advances in the understanding of brown spider venoms: From the biology of spiders to the molecular mechanisms of toxins.Toxicon. 2014 Jun;83:91-120. doi: 10.1016/j.toxicon.2014.02.023. Epub 2014 Mar 11. Toxicon. 2014. PMID: 24631373 Review.
Cited by
-
Advanced Situation with Recombinant Toxins: Diversity, Production and Application Purposes.Int J Mol Sci. 2023 Feb 27;24(5):4630. doi: 10.3390/ijms24054630. Int J Mol Sci. 2023. PMID: 36902061 Free PMC article. Review.
-
Venom-gland transcriptomics and venom proteomics of the Tibellus oblongus spider.Sci Data. 2023 Nov 22;10(1):820. doi: 10.1038/s41597-023-02703-0. Sci Data. 2023. PMID: 37993463 Free PMC article.
-
Spider-Venom Peptides: Structure, Bioactivity, Strategy, and Research Applications.Molecules. 2023 Dec 20;29(1):35. doi: 10.3390/molecules29010035. Molecules. 2023. PMID: 38202621 Free PMC article. Review.
-
Composition and toxicity of venom produced by araneophagous white-tailed spiders (Lamponidae: Lampona sp.).Sci Rep. 2022 Dec 14;12(1):21597. doi: 10.1038/s41598-022-24694-5. Sci Rep. 2022. PMID: 36517485 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

