Genomic Properties and Temporal Analysis of the Interaction of an Invasive Escherichia albertii With Epithelial Cells
- PMID: 33392102
- PMCID: PMC7772469
- DOI: 10.3389/fcimb.2020.571088
Genomic Properties and Temporal Analysis of the Interaction of an Invasive Escherichia albertii With Epithelial Cells
Abstract
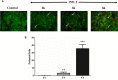
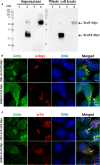
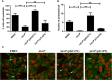
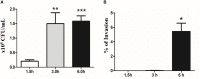
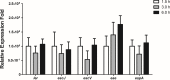
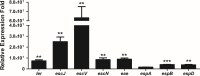
Diarrhea is one of the main causes of infant mortality worldwide, mainly in the developing world. Among the various etiologic agents, Escherichia albertii is emerging as an important human enteropathogen. E. albertii promote attaching and effacing (AE) lesions due to the presence of the locus of enterocyte effacement (LEE) that encodes a type three secretion system (T3SS), the afimbrial adhesin intimin and its translocated receptor, Tir, and several effector proteins. We previously showed that E. albertii strain 1551-2 invades several epithelial cell lineages by a process that is dependent on the intimin-Tir interaction. To understand the contribution of T3SS-dependent effectors present in E. albertii 1551-2 during the invasion process, we performed a genetic analysis of the LEE and non-LEE genes and evaluated the expression of the LEE operons in various stages of bacterial interaction with differentiated intestinal Caco-2 cells. The kinetics of the ability of the 1551-2 strain to colonize and form AE lesions was also investigated in epithelial HeLa cells. We showed that the LEE expression was constant during the early stages of infection but increased at least 4-fold during bacterial persistence in the intracellular compartment. An in silico analysis indicated the presence of a new tccP/espFU subtype, named tccP3. We found that the encoded protein colocalizes with Tir and polymerized F-actin during the infection process in vitro. Moreover, assays performed with Nck null cells demonstrated that the 1551-2 strain can trigger F-actin polymerization in an Nck-independent pathway, despite the fact that TccP3 is not required for this phenotype. Our study highlights the importance of the T3SS during the invasion process and for the maintenance of E. albertii 1551-2 inside the cells. In addition, this work may help to elucidate the versatility of the T3SS for AE pathogens, which are usually considered extracellular and rarely reach the intracellular environment.
Keywords: Escherichia albertii; Tir cytoskeleton-coupling protein/EspFu; attaching and effacing lesion; diarrhea; invasion; locus of enterocyte effacement; pathogenicity; type three secretion system.
Copyright © 2020 Romão, Martins, Hernandes, Ooka, Santos, Yamamoto, Bonfim-Melo, Jones, Hayashi, Elias, Sperandio and Gomes.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
EspFu-Mediated Actin Assembly Enhances Enteropathogenic Escherichia coli Adherence and Activates Host Cell Inflammatory Signaling Pathways.mBio. 2020 Apr 14;11(2):e00617-20. doi: 10.1128/mBio.00617-20. mBio. 2020. PMID: 32291304 Free PMC article.
-
Nck adaptors, besides promoting N-WASP mediated actin-nucleation activity at pedestals, influence the cellular levels of enteropathogenic Escherichia coli Tir effector.Cell Adh Migr. 2014;8(4):404-17. doi: 10.4161/19336918.2014.969993. Cell Adh Migr. 2014. PMID: 25482634 Free PMC article.
-
Attaching and effacing (A/E) lesion formation by enteropathogenic E. coli on human intestinal mucosa is dependent on non-LEE effectors.PLoS Pathog. 2017 Oct 30;13(10):e1006706. doi: 10.1371/journal.ppat.1006706. eCollection 2017 Oct. PLoS Pathog. 2017. PMID: 29084270 Free PMC article.
-
Interkingdom Chemical Signaling in Enterohemorrhagic Escherichia coli O157:H7.Adv Exp Med Biol. 2016;874:201-13. doi: 10.1007/978-3-319-20215-0_9. Adv Exp Med Biol. 2016. PMID: 26589220 Review.
-
Locus of enterocyte effacement: a pathogenicity island involved in the virulence of enteropathogenic and enterohemorragic Escherichia coli subjected to a complex network of gene regulation.Biomed Res Int. 2015;2015:534738. doi: 10.1155/2015/534738. Epub 2015 Feb 2. Biomed Res Int. 2015. PMID: 25710006 Free PMC article. Review.
Cited by
-
ACE2, ACE, DPPIV, PREP and CAT L enzymatic activities in COVID-19: imbalance of ACE2/ACE ratio and potential RAAS dysregulation in severe cases.Inflamm Res. 2023 Aug;72(8):1719-1731. doi: 10.1007/s00011-023-01775-3. Epub 2023 Aug 3. Inflamm Res. 2023. PMID: 37537367
-
Expression of the locus of enterocyte effacement genes during the invasion process of the atypical enteropathogenic Escherichia coli 1711-4 strain of serotype O51:H40.Microbiol Spectr. 2024 Oct 3;12(10):e0030424. doi: 10.1128/spectrum.00304-24. Epub 2024 Aug 27. Microbiol Spectr. 2024. PMID: 39189752 Free PMC article.
References
-
- Bladt F., Aippersbach E., Gelkop S., Strasser G. A., Nash P., Tafuri A., et al. (2003). The murine Nck SH2/SH3 adaptors are important for the development of mesoderm-derived embryonic structures and for regulating the cellular actin network. Mol. Cell. Biol. 23, 4586–4597. 10.1128/mcb.23.13.4586-4597.2003 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

