RNA-binding protein HuR suppresses senescence through Atg7 mediated autophagy activation in diabetic intervertebral disc degeneration
- PMID: 33372336
- PMCID: PMC7848958
- DOI: 10.1111/cpr.12975
RNA-binding protein HuR suppresses senescence through Atg7 mediated autophagy activation in diabetic intervertebral disc degeneration
Abstract
Objectives: Diabetes is a risk factor for intervertebral disc degeneration (IVDD). Studies have demonstrated that diabetes may affect IVDD through transcriptional regulation; however, whether post-transcriptional regulation is involved in diabetic IVDD (DB-IVDD) is still unknown. This study was performed to illustrate the role of HuR, an RNA-binding protein, in DB-IVDD development and its mechanism.
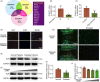
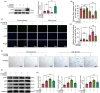
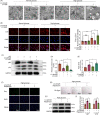
Materials and methods: The expression of HuR was evaluated in nucleus pulposus (NP) tissues from diabetic IVDD patients and in high glucose-treated NP cells. Senescence and autophagy were assessed in HuR over-expressing and downregulation NP cells. The mRNAs that were regulated by HuR were screened, and immunoprecipitation was applied to confirm the regulation of HuR on targeted mRNAs.
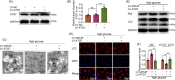
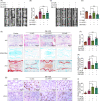
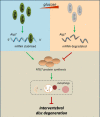
Results: The results showed that the expression of HuR was decreased in diabetic NP tissues and high glucose-treated NP cells. Downregulation of HuR may lead to increased senescence in high glucose-treated NP cells, while autophagy activation attenuates senescence in HuR deficient NP cells. Mechanistic study showed that HuR prompted Atg7 mRNA stability via binding to the AU-rich elements. Furthermore, overexpression of Atg7, but not HuR, may ameliorate DB-IVDD in rats in vivo.
Conclusions: In conclusion, HuR may suppress senescence through autophagy activation via stabilizing Atg7 in diabetic NP cells; while Atg7, but not HuR, may serve as a potential therapeutic target for DB-IVDD.
Keywords: Atg7; HuR; autophagy; diabetes; intervertebral disc degeneration; senescence.
© 2020 The Authors. Cell Proliferation Published by John Wiley & Sons Ltd.
Conflict of interest statement
There is no conflict of interest between the authors.
Figures


Similar articles
-
TFEB protects nucleus pulposus cells against apoptosis and senescence via restoring autophagic flux.Osteoarthritis Cartilage. 2019 Feb;27(2):347-357. doi: 10.1016/j.joca.2018.10.011. Epub 2018 Nov 8. Osteoarthritis Cartilage. 2019. PMID: 30414849
-
The therapeutic effect of TBK1 in intervertebral disc degeneration via coordinating selective autophagy and autophagic functions.J Adv Res. 2020 Aug 24;30:1-13. doi: 10.1016/j.jare.2020.08.011. eCollection 2021 May. J Adv Res. 2020. PMID: 34026282 Free PMC article.
-
Tbxt alleviates senescence and apoptosis of nucleus pulposus cells through Atg7-mediated autophagy activation during intervertebral disk degeneration.Am J Physiol Cell Physiol. 2024 Aug 1;327(2):C237-C253. doi: 10.1152/ajpcell.00126.2024. Epub 2024 Jun 10. Am J Physiol Cell Physiol. 2024. PMID: 38853649
-
Metformin prevents the onset and progression of intervertebral disc degeneration: New insights and potential mechanisms (Review).Int J Mol Med. 2024 Aug;54(2):71. doi: 10.3892/ijmm.2024.5395. Epub 2024 Jul 4. Int J Mol Med. 2024. PMID: 38963023 Free PMC article. Review.
-
Cellular senescence - Molecular mechanisms of intervertebral disc degeneration from an immune perspective.Biomed Pharmacother. 2023 Jun;162:114711. doi: 10.1016/j.biopha.2023.114711. Epub 2023 Apr 20. Biomed Pharmacother. 2023. PMID: 37084562 Review.
Cited by
-
Eicosapentaenoic Acid-Induced Autophagy Attenuates Intervertebral Disc Degeneration by Suppressing Endoplasmic Reticulum Stress, Extracellular Matrix Degradation, and Apoptosis.Front Cell Dev Biol. 2021 Nov 4;9:745621. doi: 10.3389/fcell.2021.745621. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34805156 Free PMC article.
-
BMP7 ameliorates intervertebral disc degeneration in type 1 diabetic rats by inhibiting pyroptosis of nucleus pulposus cells and NLRP3 inflammasome activity.Mol Med. 2023 Mar 1;29(1):30. doi: 10.1186/s10020-023-00623-8. Mol Med. 2023. PMID: 36858954 Free PMC article.
-
Emerging roles of RNA binding proteins in intervertebral disc degeneration and osteoarthritis.Orthop Surg. 2023 Dec;15(12):3015-3025. doi: 10.1111/os.13851. Epub 2023 Oct 6. Orthop Surg. 2023. PMID: 37803912 Free PMC article. Review.
-
DNA and protein methyltransferases inhibition by adenosine dialdehyde reduces the proliferation and migration of breast and lung cancer cells by downregulating autophagy.PLoS One. 2023 Jul 28;18(7):e0288791. doi: 10.1371/journal.pone.0288791. eCollection 2023. PLoS One. 2023. PMID: 37506102 Free PMC article.
-
Regulation of autophagy gene expression and its implications in cancer.J Cell Sci. 2023 May 15;136(10):jcs260631. doi: 10.1242/jcs.260631. Epub 2023 May 18. J Cell Sci. 2023. PMID: 37199330 Free PMC article. Review.
References
-
- Hoy D, Bain C, Williams G, et al. A systematic review of the global prevalence of low back pain. Arthritis Rheum. 2012;64(6):2028‐2037. - PubMed
-
- Jensen CE, Riis A, Petersen KD, Jensen MB, Pedersen KM. Economic evaluation of an implementation strategy for the management of low back pain in general practice. Pain. 2017;158(5):891‐899. - PubMed
-
- Martin BI, Deyo RA, Mirza SK, et al. Expenditures and health status among adults with back and neck problems. JAMA. 2008;299(6):656‐664. - PubMed
MeSH terms
Substances
Grants and funding
- 2017KY463/Zhejiang Medical Science Foundation
- 2020KY190/Zhejiang Medical Science Foundation
- ZY2019014/Wenzhou Science and Technology Bureau Foundation
- 81871806/National Natural Science Foundation of China
- 81902243/National Natural Science Foundation of China
- 81972094/National Natural Science Foundation of China
- LGF20H060013/Zhejiang Provincial Natural Science Foundation of China
- LGF21H060011/Zhejiang Provincial Natural Science Foundation of China
- LQ19H060004/Zhejiang Provincial Natural Science Foundation of China
- LY17H250002/Zhejiang Provincial Natural Science Foundation of China
- LY18H060012/Zhejiang Provincial Natural Science Foundation of China
- 18331213/Lin He's New Medicine and Clinical Translation Academician Workstation Research Fund
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

