Targeting host cell proteases as a potential treatment strategy to limit the spread of SARS-CoV-2 in the respiratory tract
- PMID: 33369210
- PMCID: PMC7758277
- DOI: 10.1002/prp2.698
Targeting host cell proteases as a potential treatment strategy to limit the spread of SARS-CoV-2 in the respiratory tract
Abstract
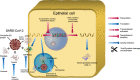
As the death toll of Coronavirus disease 19 (COVID-19) continues to rise worldwide, it is imperative to explore novel molecular mechanisms for targeting SARS-CoV-2. Rather than looking for drugs that directly interact with key viral proteins inhibiting its replication, an alternative and possibly add-on approach is to dismantle the host cell machinery that enables the virus to infect the host cell and spread from one cell to another. Excellent examples of such machinery are host cell proteases whose role in viral pathogenesis has been demonstrated in numerous coronaviruses. In this review, we propose two therapeutic modalities to tackle SARS-CoV-2 infections; the first is to transcriptionally modulate the expression of cellular proteases and their endogenous inhibitors and the second is to directly inhibit their enzymatic activity. We present a nonexhaustive collection of clinically investigated drugs that act by one of these mechanisms and thus represent promising candidates for preclinical in vitro testing and hopefully clinical testing in COVID-19 patients.
Keywords: COVID-19; SARS-CoV-2; adjunctive therapy; clinical trial; proteases.
© 2020 The Authors. Pharmacology Research & Perspectives published by John Wiley & Sons Ltd, British Pharmacological Society and American Society for Pharmacology and Experimental Therapeutics.
Conflict of interest statement
None.
Figures

Similar articles
-
Host Cell Proteases Involved in Human Respiratory Viral Infections and Their Inhibitors: A Review.Viruses. 2024 Jun 19;16(6):984. doi: 10.3390/v16060984. Viruses. 2024. PMID: 38932275 Free PMC article. Review.
-
Host Cell Proteases Mediating SARS-CoV-2 Entry: An Overview.Curr Top Med Chem. 2022;22(21):1776-1792. doi: 10.2174/1568026622666220726122339. Curr Top Med Chem. 2022. PMID: 35894476 Review.
-
Overview of antiviral drug candidates targeting coronaviral 3C-like main proteases.FEBS J. 2021 Sep;288(17):5089-5121. doi: 10.1111/febs.15696. Epub 2021 Feb 1. FEBS J. 2021. PMID: 33400393 Review.
-
Potential of coronavirus 3C-like protease inhibitors for the development of new anti-SARS-CoV-2 drugs: Insights from structures of protease and inhibitors.Int J Antimicrob Agents. 2020 Aug;56(2):106055. doi: 10.1016/j.ijantimicag.2020.106055. Epub 2020 Jun 11. Int J Antimicrob Agents. 2020. PMID: 32534187 Free PMC article.
-
SARS-CoV-2 strategically mimics proteolytic activation of human ENaC.Elife. 2020 May 26;9:e58603. doi: 10.7554/eLife.58603. Elife. 2020. PMID: 32452762 Free PMC article.
Cited by
-
Integrin/TGF-β1 Inhibitor GLPG-0187 Blocks SARS-CoV-2 Delta and Omicron Pseudovirus Infection of Airway Epithelial Cells In Vitro, Which Could Attenuate Disease Severity.Pharmaceuticals (Basel). 2022 May 17;15(5):618. doi: 10.3390/ph15050618. Pharmaceuticals (Basel). 2022. PMID: 35631444 Free PMC article.
-
Recent Advances on Targeting Proteases for Antiviral Development.Viruses. 2024 Feb 27;16(3):366. doi: 10.3390/v16030366. Viruses. 2024. PMID: 38543732 Free PMC article. Review.
-
COVID-19-Associated acute respiratory distress syndrome (CARDS): Mechanistic insights on therapeutic intervention and emerging trends.Int Immunopharmacol. 2021 Dec;101(Pt A):108328. doi: 10.1016/j.intimp.2021.108328. Epub 2021 Nov 3. Int Immunopharmacol. 2021. PMID: 34768236 Free PMC article.
-
Hyperinflammatory Response in COVID-19: A Systematic Review.Viruses. 2023 Feb 16;15(2):553. doi: 10.3390/v15020553. Viruses. 2023. PMID: 36851766 Free PMC article. Review.
-
Coordination chemistry suggests that independently observed benefits of metformin and Zn2+ against COVID-19 are not independent.Biometals. 2024 Aug;37(4):983-1022. doi: 10.1007/s10534-024-00590-5. Epub 2024 Apr 5. Biometals. 2024. PMID: 38578560 Free PMC article.
References
-
- Coronavirus disease (COVID‐19) outbreak. World Health Organization (WHO) https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019.
-
- Hariri L, Hardin CC. Covid‐19, angiogenesis, and ARDS endotypes. N Engl J Med. 2020;383:182‐183. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

