Mono-Alkylated Ligands Based on Pyrazole and Triazole Derivatives Tested Against Fusarium oxysporum f. sp. albedinis: Synthesis, Characterization, DFT, and Phytase Binding Site Identification Using Blind Docking/Virtual Screening for Potent Fophy Inhibitors
- PMID: 33363103
- PMCID: PMC7759635
- DOI: 10.3389/fchem.2020.559262
Mono-Alkylated Ligands Based on Pyrazole and Triazole Derivatives Tested Against Fusarium oxysporum f. sp. albedinis: Synthesis, Characterization, DFT, and Phytase Binding Site Identification Using Blind Docking/Virtual Screening for Potent Fophy Inhibitors
Abstract
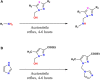
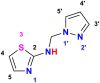
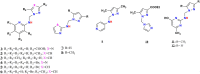
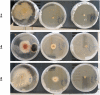
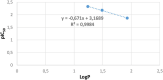
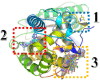
Twelve recent compounds, incorporating several heterocyclic moieties such as pyrazole, thiazole, triazole, and benzotriazole, made in excellent yield up to 37-99.6%. They were tested against Fusarium oxysporum f. sp. albedinis fungi (Bayoud disease), where the best results are for compounds 2, 4, and 5 with IC50 = 18.8-54.4 μg/mL. Density functional theory (DFT) study presented their molecular reactivity, while the docking simulations to describe the synergies between the trained compounds of dataset containing all the tested compounds (57 molecules) and F. oxysporum phytase domain (Fophy) enzyme as biological target. By comparing the results of the docking studies for the Fophy protein, it is found that compound 5 has the best affinity followed by compounds 2 and 4, so there is good agreement with the experimental results where their IC50 values are in the following order: 74.28 (5) < 150 (2) < 214.10 (4), using Blind docking/virtual screening of the homology modeled protein and two different tools as Autodock Vina and Dockthor web tool that gave us predicted sites for further antifungal drug design.
Keywords: DFT; Fusarium oxysporum; antifungal; docking; pyrazole; triazole.
Copyright © 2020 Kaddouri, Abrigach, Ouahhoud, Benabbes, El Kodadi, Alsalme, Al-Zaqri, Warad and Touzani.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



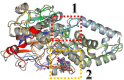
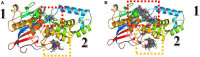
Similar articles
-
Synthesis, characterization, reaction mechanism prediction and biological study of mono, bis and tetrakis pyrazole derivatives against Fusarium oxysporum f. sp. Albedinis with conceptual DFT and ligand-protein docking studies.Bioorg Chem. 2021 May;110:104696. doi: 10.1016/j.bioorg.2021.104696. Epub 2021 Feb 8. Bioorg Chem. 2021. PMID: 33652343
-
An Insight into All Tested Small Molecules against Fusarium oxysporum f. sp. Albedinis: A Comparative Review.Molecules. 2022 Apr 22;27(9):2698. doi: 10.3390/molecules27092698. Molecules. 2022. PMID: 35566050 Free PMC article. Review.
-
Fot 1 insertions in the Fusarium oxysporum f. sp. albedinis genome provide diagnostic PCR targets for detection of the date palm pathogen.Appl Environ Microbiol. 1998 Feb;64(2):633-6. doi: 10.1128/AEM.64.2.633-636.1998. Appl Environ Microbiol. 1998. PMID: 9464402 Free PMC article.
-
New thiazole, pyridine and pyrazole derivatives as antioxidant candidates: synthesis, DFT calculations and molecular docking study.Heliyon. 2020 Jan 9;6(1):e03185. doi: 10.1016/j.heliyon.2020.e03185. eCollection 2020 Jan. Heliyon. 2020. PMID: 31956713 Free PMC article.
-
In vitro screening, homology modeling and molecular docking studies of some pyrazole and imidazole derivatives.Biomed Pharmacother. 2018 Jul;103:653-661. doi: 10.1016/j.biopha.2018.04.061. Epub 2018 Apr 24. Biomed Pharmacother. 2018. PMID: 29679907
Cited by
-
New N-Alkylated Heterocyclic Compounds as Prospective NDM1 Inhibitors: Investigation of In Vitro and In Silico Properties.Pharmaceuticals (Basel). 2022 Jun 28;15(7):803. doi: 10.3390/ph15070803. Pharmaceuticals (Basel). 2022. PMID: 35890102 Free PMC article.
-
Coordination Complexes Built from a Ditopic Triazole-Pyrazole Ligand with Antibacterial and Antifungal Performances.Molecules. 2023 Sep 25;28(19):6801. doi: 10.3390/molecules28196801. Molecules. 2023. PMID: 37836644 Free PMC article.
-
Novel family of bis-pyrazole coordination complexes as potent antibacterial and antifungal agents.RSC Adv. 2022 Jun 15;12(28):17755-17764. doi: 10.1039/d2ra03414j. eCollection 2022 Jun 14. RSC Adv. 2022. PMID: 35765319 Free PMC article.
-
Pyrazole, imidazole and triazole: In silico, docking and ADMET studies against SARS-CoV-2.Mater Today Proc. 2023;72:3686-3695. doi: 10.1016/j.matpr.2022.09.060. Epub 2022 Sep 9. Mater Today Proc. 2023. PMID: 36101672 Free PMC article.
-
Synthesis, Crystal Structures, Genotoxicity, and Antifungal and Antibacterial Studies of Ni(II) and Cd(II) Pyrazole Amide Coordination Complexes.Molecules. 2024 Mar 6;29(5):1186. doi: 10.3390/molecules29051186. Molecules. 2024. PMID: 38474698 Free PMC article.
References
-
- Abrigach F., Karzazi Y., Benabbed R., Elyoubi M., Khoutoul M., Taibi N., et al. (2017). Synthesis, biological screening, POM and 3D-QSAR analyses of some novel pyrazolic compounds. Med. Chem. Res. 26, 1784–1795. 10.1007/s00044-017-1888-8 - DOI
-
- Abrigach F., Khoutoul M., Merghache S., Oussaid A., Lamsayah M., Zarrouk A., et al. (2014). Antioxidant activities of N-((3,5-dimethyl-1H-pyrazol-1-yl)methyl)pyridin-4-amine derivatives. Der. Pharma. Chem. 6, 280–285.
-
- Abrigach F., Rokni Y., Takfaoui A., Khoutoul M., Doucet H. (2018). In vitro screening, homology modeling and molecular docking studies of some pyrazole and imidazole derivatives biomedicine & pharmacotherapy in vitro screening, homology modeling and molecular docking studies of some pyrazole and imidazole derivatives. Biomed. Pharmacother. 103, 653–661. 10.1016/j.biopha.2018.04.061 - DOI - PubMed
LinkOut - more resources
Full Text Sources

