Single-Cell Transcriptomics Reveals a Heterogeneous Cellular Response to BK Virus Infection
- PMID: 33361432
- PMCID: PMC8094954
- DOI: 10.1128/JVI.02237-20
Single-Cell Transcriptomics Reveals a Heterogeneous Cellular Response to BK Virus Infection
Abstract
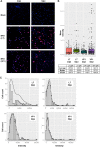
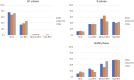
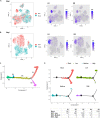
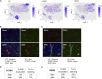
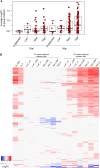
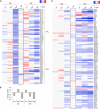
BK virus (BKV) is a human polyomavirus that is generally harmless but can cause devastating disease in immunosuppressed individuals. BKV infection of renal cells is a common problem for kidney transplant patients undergoing immunosuppressive therapy. In cultured primary human renal proximal tubule epithelial (RPTE) cells, BKV undergoes a productive infection. The BKV-encoded large T antigen (LT) induces cell cycle entry, resulting in the upregulation of numerous genes associated with cell proliferation. Consistently, microarray and transcriptome sequencing (RNA-seq) experiments performed on bulk infected cell populations identified several proliferation-related pathways that are upregulated by BKV. These studies revealed few genes that are downregulated. In this study, we analyzed viral and cellular transcripts in single mock- or BKV-infected cells. We found that the levels of viral mRNAs vary widely among infected cells, resulting in different levels of LT and viral capsid protein expression. Cells expressing the highest levels of viral transcripts account for approximately 20% of the culture and have a gene expression pattern that is distinct from that of cells expressing lower levels of viral mRNAs. Surprisingly, cells expressing low levels of viral mRNA do not progress with time to high expression, suggesting that the two cellular responses are determined prior to or shortly following infection. Finally, comparison of cellular gene expression patterns of cells expressing high levels of viral mRNA with those of mock-infected cells or cells expressing low levels of viral mRNA revealed previously unidentified pathways that are downregulated by BKV. Among these are pathways associated with drug metabolism and detoxification, tumor necrosis factor (TNF) signaling, energy metabolism, and translation.IMPORTANCE The outcome of viral infection is determined by the ability of the virus to redirect cellular systems toward progeny production countered by the ability of the cell to block these viral actions. Thus, an infected culture consists of thousands of cells, each fighting its own individual battle. Bulk measurements, such as PCR or RNA-seq, measure the average of these individual responses to infection. Single-cell transcriptomics provides a window to the one-on-one battle between BKV and each cell. Our studies reveal that only a minority of infected cells are overwhelmed by the virus and produce large amounts of BKV mRNAs and proteins, while the infection appears to be restricted in the remaining cells. Correlation of viral transcript levels with cellular gene expression patterns reveals pathways manipulated by BKV that may play a role in limiting infection.
Keywords: BKV; polyomavirus; single-cell transcriptomics.
Copyright © 2021 American Society for Microbiology.
Figures





Similar articles
-
Cultured Renal Proximal Tubular Epithelial Cells Resemble a Stressed/Damaged Kidney While Supporting BK Virus Infection.J Virol. 2023 May 31;97(5):e0034323. doi: 10.1128/jvi.00343-23. Epub 2023 May 11. J Virol. 2023. PMID: 37166336 Free PMC article.
-
Polyomavirus BK-encoded microRNA suppresses autoregulation of viral replication.Biochem Biophys Res Commun. 2014 May 9;447(3):543-9. doi: 10.1016/j.bbrc.2014.04.030. Epub 2014 Apr 13. Biochem Biophys Res Commun. 2014. PMID: 24735545
-
Cellular and viral miRNA expression in polyomavirus BK infection.Transpl Infect Dis. 2019 Oct;21(5):e13159. doi: 10.1111/tid.13159. Epub 2019 Aug 29. Transpl Infect Dis. 2019. PMID: 31410940
-
BK virus large T and VP-1 expression in infected human renal allografts.Nephrol Dial Transplant. 2008 Dec;23(12):3752-61. doi: 10.1093/ndt/gfn470. Epub 2008 Sep 9. Nephrol Dial Transplant. 2008. PMID: 18784088 Free PMC article. Review.
-
The polyomavirus puzzle: is host immune response beneficial in controlling BK virus after adult hematopoietic cell transplantion?Transpl Infect Dis. 2014 Aug;16(4):521-31. doi: 10.1111/tid.12233. Epub 2014 May 19. Transpl Infect Dis. 2014. PMID: 24834968 Free PMC article. Review.
Cited by
-
Human genes with relative synonymous codon usage analogous to that of polyomaviruses are involved in the mechanism of polyomavirus nephropathy.Front Cell Infect Microbiol. 2022 Sep 8;12:992201. doi: 10.3389/fcimb.2022.992201. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36159639 Free PMC article.
-
Lessons from Polyomavirus Immunofluorescence Staining of Urinary Decoy Cells.Life (Basel). 2023 Jul 7;13(7):1526. doi: 10.3390/life13071526. Life (Basel). 2023. PMID: 37511901 Free PMC article.
-
Single-Cell Transcriptome Identifies the Renal Cell Type Tropism of Human BK Polyomavirus.Int J Mol Sci. 2023 Jan 10;24(2):1330. doi: 10.3390/ijms24021330. Int J Mol Sci. 2023. PMID: 36674845 Free PMC article.
-
Single-Cell, High-Content Microscopy Analysis of BK Polyomavirus Infection.Microbiol Spectr. 2023 Jun 15;11(3):e0087323. doi: 10.1128/spectrum.00873-23. Epub 2023 May 8. Microbiol Spectr. 2023. PMID: 37154756 Free PMC article.
-
Cultured Renal Proximal Tubular Epithelial Cells Resemble a Stressed/Damaged Kidney While Supporting BK Virus Infection.J Virol. 2023 May 31;97(5):e0034323. doi: 10.1128/jvi.00343-23. Epub 2023 May 11. J Virol. 2023. PMID: 37166336 Free PMC article.
References
-
- Antonsson A, Pawlita M, Feltkamp MCW, Bouwes Bavinck JN, Euvrard S, Harwood CA, Naldi L, Nindl I, Proby CM, Neale RE, Waterboer T. 2013. Longitudinal study of seroprevalence and serostability of the human polyomaviruses JCV and BKV in organ transplant recipients. J Med Virol 85:327–335. doi:10.1002/jmv.23472. - DOI - PubMed
-
- Koskenvuo M, Dumoulin A, Lautenschlager I, Auvinen E, Mannonen L, Anttila VJ, Jahnukainen K, Saarinen-Pihkala UM, Hirsch HH. 2013. BK polyomavirus-associated hemorrhagic cystitis among pediatric allogeneic bone marrow transplant recipients: treatment response and evidence for nosocomial transmission. J Clin Virol 56:77–81. doi:10.1016/j.jcv.2012.09.003. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

