c-Jun N-Terminal Kinase Inhibitors as Potential Leads for New Therapeutics for Alzheimer's Diseases
- PMID: 33352989
- PMCID: PMC7765872
- DOI: 10.3390/ijms21249677
c-Jun N-Terminal Kinase Inhibitors as Potential Leads for New Therapeutics for Alzheimer's Diseases
Abstract
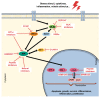
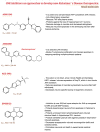
Alzheimer's Disease (AD) is becoming more prevalent as the population lives longer. For individuals over 60 years of age, the prevalence of AD is estimated at 40.19% across the world. Regarding the cognitive decline caused by the disease, mitogen-activated protein kinases (MAPK) pathways such as the c-Jun N-terminal kinase (JNK) pathway are involved in the progressive loss of neurons and synapses, brain atrophy, and augmentation of the brain ventricles, being activated by synaptic dysfunction, oxidative stress, and excitotoxicity. Nowadays, AD symptoms are manageable, but the disease itself remains incurable, thus the inhibition of JNK3 has been explored as a possible therapeutic target, considering that JNK is best known for its involvement in propagating pro-apoptotic signals. This review aims to present biological aspects of JNK, focusing on JNK3 and how it relates to AD. It was also explored the recent development of inhibitors that could be used in AD treatment since several drugs/compounds in phase III clinical trials failed. General aspects of the MAPK family, therapeutic targets, and experimental treatment in models are described and discussed throughout this review.
Keywords: brain diseases; c-Jun N-terminal kinase (JNK); kinase inhibitors; therapeutic targets.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results
Figures



Similar articles
-
Activation and redistribution of c-jun N-terminal kinase/stress activated protein kinase in degenerating neurons in Alzheimer's disease.J Neurochem. 2001 Jan;76(2):435-41. doi: 10.1046/j.1471-4159.2001.00046.x. J Neurochem. 2001. PMID: 11208906
-
c-Jun N-terminal kinase inhibitors: a patent review (2010 - 2014).Expert Opin Ther Pat. 2015;25(8):849-72. doi: 10.1517/13543776.2015.1039984. Epub 2015 May 19. Expert Opin Ther Pat. 2015. PMID: 25991433 Review.
-
c-Jun N-Terminal Kinases in Alzheimer's Disease: A Possible Target for the Modulation of the Earliest Alterations.J Alzheimers Dis. 2021;82(s1):S127-S139. doi: 10.3233/JAD-201053. J Alzheimers Dis. 2021. PMID: 33216036 Review.
-
c-Jun N-terminal kinase has a key role in Alzheimer disease synaptic dysfunction in vivo.Cell Death Dis. 2014 Jan 23;5(1):e1019. doi: 10.1038/cddis.2013.559. Cell Death Dis. 2014. PMID: 24457963 Free PMC article.
-
JNK: a stress-activated protein kinase therapeutic strategies and involvement in Alzheimer's and various neurodegenerative abnormalities.J Mol Neurosci. 2011 Mar;43(3):376-90. doi: 10.1007/s12031-010-9454-6. Epub 2010 Sep 28. J Mol Neurosci. 2011. PMID: 20878262 Review.
Cited by
-
Novel Tryptanthrin Derivatives with Selectivity as c-Jun N-Terminal Kinase (JNK) 3 Inhibitors.Molecules. 2023 Jun 16;28(12):4806. doi: 10.3390/molecules28124806. Molecules. 2023. PMID: 37375361 Free PMC article.
-
Neuroprotective Effect of Luteolin-7-O-Glucoside against 6-OHDA-Induced Damage in Undifferentiated and RA-Differentiated SH-SY5Y Cells.Int J Mol Sci. 2022 Mar 8;23(6):2914. doi: 10.3390/ijms23062914. Int J Mol Sci. 2022. PMID: 35328335 Free PMC article.
-
Structural Study of Selectivity Mechanisms for JNK3 and p38α with Indazole Scaffold Probing Compounds.Res Sq [Preprint]. 2024 Aug 7:rs.3.rs-4730282. doi: 10.21203/rs.3.rs-4730282/v1. Res Sq. 2024. PMID: 39149466 Free PMC article. Preprint.
-
JNK3 inhibitors as promising pharmaceuticals with neuroprotective properties.Cell Adh Migr. 2024 Dec;18(1):1-11. doi: 10.1080/19336918.2024.2316576. Epub 2024 Feb 15. Cell Adh Migr. 2024. PMID: 38357988 Free PMC article. Review.
-
The Role of the Dysregulated JNK Signaling Pathway in the Pathogenesis of Human Diseases and Its Potential Therapeutic Strategies: A Comprehensive Review.Biomolecules. 2024 Feb 19;14(2):243. doi: 10.3390/biom14020243. Biomolecules. 2024. PMID: 38397480 Free PMC article. Review.
References
-
- Alzheimer A. Über eine eigenartige Erkrankung der Hirnrinde. Allgemeine Zeitschrift fur Psychiatrie und Psychisch-gerichttliche Medizin. 1907;64:146–148.
-
- Nichols E., Szoeke C.E.I., Vollset S.E., Abbasi N., Abd-Allah F., Abdela J., Aichour M.T.E., Akinyemi R.O., Alahdab F., Asgedom S.W., et al. Global, regional, and national burden of Alzheimer’s disease and other dementias, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18:88–106. doi: 10.1016/S1474-4422(18)30403-4. - DOI - PMC - PubMed
-
- Alzheimer’s Association Alzheimer’s Disease Facts and Figures. Alzheimer’s Dementia. 2019;15:321–387. doi: 10.1016/j.jalz.2019.01.010. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

