Downregulation of Glutamine Synthetase, not glutaminolysis, is responsible for glutamine addiction in Notch1-driven acute lymphoblastic leukemia
- PMID: 33314742
- PMCID: PMC8096784
- DOI: 10.1002/1878-0261.12877
Downregulation of Glutamine Synthetase, not glutaminolysis, is responsible for glutamine addiction in Notch1-driven acute lymphoblastic leukemia
Abstract
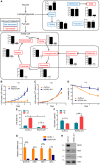
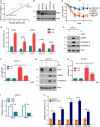
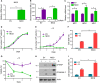
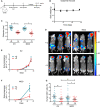
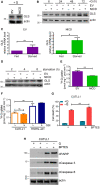
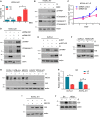
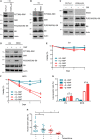
The cellular receptor Notch1 is a central regulator of T-cell development, and as a consequence, Notch1 pathway appears upregulated in > 65% of the cases of T-cell acute lymphoblastic leukemia (T-ALL). However, strategies targeting Notch1 signaling render only modest results in the clinic due to treatment resistance and severe side effects. While many investigations reported the different aspects of tumor cell growth and leukemia progression controlled by Notch1, less is known regarding the modifications of cellular metabolism induced by Notch1 upregulation in T-ALL. Previously, glutaminolysis inhibition has been proposed to synergize with anti-Notch therapies in T-ALL models. In this work, we report that Notch1 upregulation in T-ALL induced a change in the metabolism of the important amino acid glutamine, preventing glutamine synthesis through the downregulation of glutamine synthetase (GS). Downregulation of GS was responsible for glutamine addiction in Notch1-driven T-ALL both in vitro and in vivo. Our results also confirmed an increase in glutaminolysis mediated by Notch1. Increased glutaminolysis resulted in the activation of the mammalian target of rapamycin complex 1 (mTORC1) pathway, a central controller of cell growth. However, glutaminolysis did not play any role in Notch1-induced glutamine addiction. Finally, the combined treatment targeting mTORC1 and limiting glutamine availability had a synergistic effect to induce apoptosis and to prevent Notch1-driven leukemia progression. Our results placed glutamine limitation and mTORC1 inhibition as a potential therapy against Notch1-driven leukemia.
Keywords: Notch1; T-cell acute lymphoblastic leukemia; glutamine; glutamine synthetase; mTORC1; metabolic addiction.
© 2020 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia.Nat Med. 2015 Oct;21(10):1182-9. doi: 10.1038/nm.3955. Epub 2015 Sep 21. Nat Med. 2015. PMID: 26390244 Free PMC article.
-
Inhibition of mitochondrial complex I reverses NOTCH1-driven metabolic reprogramming in T-cell acute lymphoblastic leukemia.Nat Commun. 2022 May 19;13(1):2801. doi: 10.1038/s41467-022-30396-3. Nat Commun. 2022. PMID: 35589701 Free PMC article.
-
Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia.J Clin Invest. 2008 Sep;118(9):3181-94. doi: 10.1172/JCI35090. J Clin Invest. 2008. PMID: 18677410 Free PMC article.
-
New insights into Notch1 regulation of the PI3K-AKT-mTOR1 signaling axis: targeted therapy of γ-secretase inhibitor resistant T-cell acute lymphoblastic leukemia.Cell Signal. 2014 Jan;26(1):149-61. doi: 10.1016/j.cellsig.2013.09.021. Epub 2013 Oct 16. Cell Signal. 2014. PMID: 24140475 Review.
-
The relevance of PTEN-AKT in relation to NOTCH1-directed treatment strategies in T-cell acute lymphoblastic leukemia.Haematologica. 2016 Sep;101(9):1010-7. doi: 10.3324/haematol.2016.146381. Haematologica. 2016. PMID: 27582570 Free PMC article. Review.
Cited by
-
Dietary Manipulation of Amino Acids for Cancer Therapy.Nutrients. 2023 Jun 25;15(13):2879. doi: 10.3390/nu15132879. Nutrients. 2023. PMID: 37447206 Free PMC article. Review.
-
Notch signaling pathway in cancer: from mechanistic insights to targeted therapies.Signal Transduct Target Ther. 2024 May 27;9(1):128. doi: 10.1038/s41392-024-01828-x. Signal Transduct Target Ther. 2024. PMID: 38797752 Free PMC article. Review.
-
Overcoming Glucocorticoid Resistance in Acute Lymphoblastic Leukemia: Repurposed Drugs Can Improve the Protocol.Front Oncol. 2021 Mar 11;11:617937. doi: 10.3389/fonc.2021.617937. eCollection 2021. Front Oncol. 2021. PMID: 33777761 Free PMC article. Review.
-
TIM-3/Galectin-9 interaction and glutamine metabolism in AML cell lines, HL-60 and THP-1.BMC Cancer. 2024 Jan 24;24(1):125. doi: 10.1186/s12885-024-11898-3. BMC Cancer. 2024. PMID: 38267906 Free PMC article.
-
Targeting Glutaminolysis Shows Efficacy in Both Prednisolone-Sensitive and in Metabolically Rewired Prednisolone-Resistant B-Cell Childhood Acute Lymphoblastic Leukaemia Cells.Int J Mol Sci. 2023 Feb 8;24(4):3378. doi: 10.3390/ijms24043378. Int J Mol Sci. 2023. PMID: 36834787 Free PMC article.
References
-
- Tremblay CS & Curtis DJ (2014) The clonal evolution of leukemic stem cells in T‐cell acute lymphoblastic leukemia. Curr Opin Hematol 21, 320–325. - PubMed
-
- Tzoneva G & Ferrando AA (2012) Recent advances on NOTCH signaling in T‐ALL. Curr Top Microbiol Immunol 360, 163–182. - PubMed
-
- Raivio KO & Andersson LC (1982) Glutamine requirements for purine metabolism in leukemic lymphoblasts. Leuk Res 6, 111–115. - PubMed
-
- Kitoh T, Kubota M, Takimoto T, Hashimoto H, Shimizu T, Sano H, Akiyama Y & Mikawa H (1990) Metabolic basis for differential glutamine requirements of human leukemia cell lines. J Cell Physiol 143, 150–153. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

