Intruder (DD38E), a recently evolved sibling family of DD34E/Tc1 transposons in animals
- PMID: 33303022
- PMCID: PMC7731502
- DOI: 10.1186/s13100-020-00227-7
Intruder (DD38E), a recently evolved sibling family of DD34E/Tc1 transposons in animals
Abstract
Background: A family of Tc1/mariner transposons with a characteristic DD38E triad of catalytic amino acid residues, named Intruder (IT), was previously discovered in sturgeon genomes, but their evolutionary landscapes remain largely unknown.
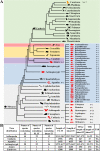
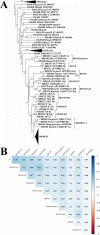
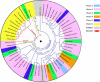
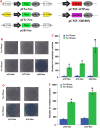
Results: Here, we comprehensively investigated the evolutionary profiles of ITs, and evaluated their cut-and-paste activities in cells. ITs exhibited a narrow taxonomic distribution pattern in the animal kingdom, with invasions into two invertebrate phyla (Arthropoda and Cnidaria) and three vertebrate lineages (Actinopterygii, Agnatha, and Anura): very similar to that of the DD36E/IC family. Some animal orders and species seem to be more hospitable to Tc1/mariner transposons, one order of Amphibia and seven Actinopterygian orders are the most common orders with horizontal transfer events and have been invaded by all four families (DD38E/IT, DD35E/TR, DD36E/IC and DD37E/TRT) of Tc1/mariner transposons, and eight Actinopterygii species were identified as the major hosts of these families. Intact ITs have a total length of 1.5-1.7 kb containing a transposase gene flanked by terminal inverted repeats (TIRs). The phylogenetic tree and sequence identity showed that IT transposases were most closely related to DD34E/Tc1. ITs have been involved in multiple events of horizontal transfer in vertebrates and have invaded most lineages recently (< 5 million years ago) based on insertion age analysis. Accordingly, ITs presented high average sequence identity (86-95%) across most vertebrate species, suggesting that some are putatively active. ITs can transpose in human HeLa cells, and the transposition efficiency of consensus TIRs was higher than that of the TIRs of natural isolates.
Conclusions: We conclude that DD38E/IT originated from DD34E/Tc1 and can be detected in two invertebrate phyla (Arthropoda and Cnidaria), and in three vertebrate lineages (Actinopterygii, Agnatha and Anura). IT has experienced multiple HT events in animals, dominated by recent amplifications in most species and has high identity among vertebrate taxa. Our reconstructed IT transposon vector designed according to the sequence from the "cat" genome showed high cut-and-paste activity. The data suggest that IT has been acquired recently and is active in many species. This study is meaningful for understanding the evolution of the Tc1/mariner superfamily members and their hosts.
Keywords: DD38E; Evolution; Horizontal transfer; Intruder; Tc1/mariner transposons.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Incomer, a DD36E family of Tc1/mariner transposons newly discovered in animals.Mob DNA. 2019 Nov 23;10:45. doi: 10.1186/s13100-019-0188-x. eCollection 2019. Mob DNA. 2019. PMID: 31788035 Free PMC article.
-
Divergent evolution profiles of DD37D and DD39D families of Tc1/mariner transposons in eukaryotes.Mol Phylogenet Evol. 2021 Aug;161:107143. doi: 10.1016/j.ympev.2021.107143. Epub 2021 Mar 10. Mol Phylogenet Evol. 2021. PMID: 33713798
-
Hiker, a new family of DNA transposons encoding transposases with DD35E motifs, displays a distinct phylogenetic relationship with most known DNA transposon families of IS630-Tc1-mariner (ITm).Mol Phylogenet Evol. 2023 Nov;188:107906. doi: 10.1016/j.ympev.2023.107906. Epub 2023 Aug 15. Mol Phylogenet Evol. 2023. PMID: 37586577
-
Mariner and the ITm Superfamily of Transposons.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0033-2014.. doi: 10.1128/microbiolspec.MDNA3-0033-2014. Microbiol Spectr. 2015. PMID: 26104691 Review.
-
Research progress of Tc1/Mariner superfamily.Yi Chuan. 2017 Jan 20;39(1):1-13. doi: 10.16288/j.yczz.16-160. Yi Chuan. 2017. PMID: 28115300 Review.
Cited by
-
Diversity and Evolution of pogo and Tc1/mariner Transposons in the Apoidea Genomes.Biology (Basel). 2021 Sep 20;10(9):940. doi: 10.3390/biology10090940. Biology (Basel). 2021. PMID: 34571816 Free PMC article.
-
Evolution of piggyBac Transposons in Apoidea.Insects. 2023 Apr 21;14(4):402. doi: 10.3390/insects14040402. Insects. 2023. PMID: 37103217 Free PMC article.
-
Horizontal Transfer and Evolutionary Profiles of Two Tc1/DD34E Transposons (ZB and SB) in Vertebrates.Genes (Basel). 2022 Nov 29;13(12):2239. doi: 10.3390/genes13122239. Genes (Basel). 2022. PMID: 36553507 Free PMC article.
-
Prokaryotic and Eukaryotic Horizontal Transfer of Sailor (DD82E), a New Superfamily of IS630-Tc1-Mariner DNA Transposons.Biology (Basel). 2021 Oct 7;10(10):1005. doi: 10.3390/biology10101005. Biology (Basel). 2021. PMID: 34681104 Free PMC article.
-
Revisiting the Tigger Transposon Evolution Revealing Extensive Involvement in the Shaping of Mammal Genomes.Biology (Basel). 2022 Jun 16;11(6):921. doi: 10.3390/biology11060921. Biology (Basel). 2022. PMID: 35741442 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

