Coexpressed subunits of dual genetic origin define a conserved supercomplex mediating essential protein import into chloroplasts
- PMID: 33273113
- PMCID: PMC7768757
- DOI: 10.1073/pnas.2014294117
Coexpressed subunits of dual genetic origin define a conserved supercomplex mediating essential protein import into chloroplasts
Abstract
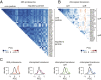
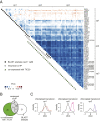
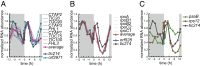
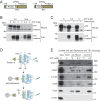
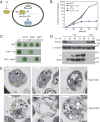
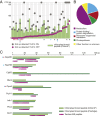
In photosynthetic eukaryotes, thousands of proteins are translated in the cytosol and imported into the chloroplast through the concerted action of two translocons-termed TOC and TIC-located in the outer and inner membranes of the chloroplast envelope, respectively. The degree to which the molecular composition of the TOC and TIC complexes is conserved over phylogenetic distances has remained controversial. Here, we combine transcriptomic, biochemical, and genetic tools in the green alga Chlamydomonas (Chlamydomonas reinhardtii) to demonstrate that, despite a lack of evident sequence conservation for some of its components, the algal TIC complex mirrors the molecular composition of a TIC complex from Arabidopsis thaliana. The Chlamydomonas TIC complex contains three nuclear-encoded subunits, Tic20, Tic56, and Tic100, and one chloroplast-encoded subunit, Tic214, and interacts with the TOC complex, as well as with several uncharacterized proteins to form a stable supercomplex (TIC-TOC), indicating that protein import across both envelope membranes is mechanistically coupled. Expression of the nuclear and chloroplast genes encoding both known and uncharacterized TIC-TOC components is highly coordinated, suggesting that a mechanism for regulating its biogenesis across compartmental boundaries must exist. Conditional repression of Tic214, the only chloroplast-encoded subunit in the TIC-TOC complex, impairs the import of chloroplast proteins with essential roles in chloroplast ribosome biogenesis and protein folding and induces a pleiotropic stress response, including several proteins involved in the chloroplast unfolded protein response. These findings underscore the functional importance of the TIC-TOC supercomplex in maintaining chloroplast proteostasis.
Keywords: Chlamydomonas reinhardtii; chloroplast gene targeting; chloroplast protein import; gene coexpression.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures

Similar articles
-
Architecture of chloroplast TOC-TIC translocon supercomplex.Nature. 2023 Mar;615(7951):349-357. doi: 10.1038/s41586-023-05744-y. Epub 2023 Jan 26. Nature. 2023. PMID: 36702157
-
Chloroplast protein import machinery and quality control.FEBS J. 2022 Nov;289(22):6908-6918. doi: 10.1111/febs.16464. Epub 2022 May 9. FEBS J. 2022. PMID: 35472255 Free PMC article. Review.
-
Structure of a TOC-TIC supercomplex spanning two chloroplast envelope membranes.Cell. 2022 Dec 8;185(25):4788-4800.e13. doi: 10.1016/j.cell.2022.10.030. Epub 2022 Nov 21. Cell. 2022. PMID: 36413996
-
The TIC complex uncovered: The alternative view on the molecular mechanism of protein translocation across the inner envelope membrane of chloroplasts.Biochim Biophys Acta. 2015 Sep;1847(9):957-67. doi: 10.1016/j.bbabio.2015.02.011. Epub 2015 Feb 16. Biochim Biophys Acta. 2015. PMID: 25689609 Review.
-
Stable megadalton TOC-TIC supercomplexes as major mediators of protein import into chloroplasts.Plant J. 2017 Oct;92(2):178-188. doi: 10.1111/tpj.13643. Epub 2017 Sep 15. Plant J. 2017. PMID: 28745032
Cited by
-
Molecular characterization, targeting and expression analysis of chloroplast and mitochondrion protein import components in Nicotiana benthamiana.Front Plant Sci. 2022 Oct 26;13:1040688. doi: 10.3389/fpls.2022.1040688. eCollection 2022. Front Plant Sci. 2022. PMID: 36388587 Free PMC article.
-
Assembly and Repair of Photosystem II in Chlamydomonas reinhardtii.Plants (Basel). 2024 Mar 12;13(6):811. doi: 10.3390/plants13060811. Plants (Basel). 2024. PMID: 38592843 Free PMC article. Review.
-
New Insights into the Chloroplast Outer Membrane Proteome and Associated Targeting Pathways.Int J Mol Sci. 2022 Jan 29;23(3):1571. doi: 10.3390/ijms23031571. Int J Mol Sci. 2022. PMID: 35163495 Free PMC article. Review.
-
A distinct class of GTP-binding proteins mediates chloroplast protein import in Rhodophyta.Proc Natl Acad Sci U S A. 2022 Aug 23;119(34):e2208277119. doi: 10.1073/pnas.2208277119. Epub 2022 Aug 15. Proc Natl Acad Sci U S A. 2022. PMID: 35969755 Free PMC article.
-
Engineered Accumulation of Bicarbonate in Plant Chloroplasts: Known Knowns and Known Unknowns.Front Plant Sci. 2021 Aug 31;12:727118. doi: 10.3389/fpls.2021.727118. eCollection 2021. Front Plant Sci. 2021. PMID: 34531888 Free PMC article. Review.
References
-
- Margulis L., Symbiotic theory of the origin of eukaryotic organelles; criteria for proof. Symp. Soc. Exp. Biol. 29, 21–38 (1975). - PubMed
-
- Chotewutmontri P., Holbrook K., Bruce B. D., Plastid protein targeting: Preprotein recognition and translocation. Int. Rev. Cell Mol. Biol. 330, 227–294 (2017). - PubMed
-
- Shi L. X., Theg S. M., The chloroplast protein import system: From algae to trees. Biochim. Biophys. Acta 1833, 314–331 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

