DEAH-Box RNA Helicases in Pre-mRNA Splicing
- PMID: 33272784
- PMCID: PMC8112905
- DOI: 10.1016/j.tibs.2020.10.006
DEAH-Box RNA Helicases in Pre-mRNA Splicing
Abstract
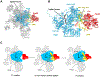
In eukaryotic cells, pre-mRNA splicing is catalyzed by the spliceosome, a highly dynamic molecular machinery that undergoes dramatic conformational and compositional rearrangements throughout the splicing cycle. These crucial rearrangements are largely driven by eight DExD/H-box RNA helicases. Interestingly, the four helicases participating in the late stages of splicing are all DEAH-box helicases that share structural similarities. This review aims to provide an overview of the structure and function of these DEAH-box helicases, including new information provided by recent cryo-electron microscopy structures of the spliceosomal complexes.
Keywords: DEAH-box RNA helicases; cryo-EM; spliceosome.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures



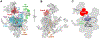


Similar articles
-
Structure, function and regulation of spliceosomal RNA helicases.Curr Opin Cell Biol. 2012 Jun;24(3):431-8. doi: 10.1016/j.ceb.2012.03.004. Epub 2012 Mar 29. Curr Opin Cell Biol. 2012. PMID: 22464735 Review.
-
RNA helicases in splicing.RNA Biol. 2013 Jan;10(1):83-95. doi: 10.4161/rna.22547. Epub 2012 Dec 10. RNA Biol. 2013. PMID: 23229095 Free PMC article. Review.
-
Structure and function of spliceosomal DEAH-box ATPases.Biol Chem. 2023 Jul 17;404(8-9):851-866. doi: 10.1515/hsz-2023-0157. Print 2023 Jul 26. Biol Chem. 2023. PMID: 37441768 Review.
-
Crystal structure of Prp16 in complex with ADP.Acta Crystallogr F Struct Biol Commun. 2023 Aug 1;79(Pt 8):200-207. doi: 10.1107/S2053230X23005721. Epub 2023 Jul 25. Acta Crystallogr F Struct Biol Commun. 2023. PMID: 37548918 Free PMC article.
-
Contribution of DEAH-box protein DHX16 in human pre-mRNA splicing.Biochem J. 2010 Jul 1;429(1):25-32. doi: 10.1042/BJ20100266. Biochem J. 2010. PMID: 20423332 Free PMC article.
Cited by
-
Porcine reproductive and respiratory syndrome virus degrades DDX10 via SQSTM1/p62-dependent selective autophagy to antagonize its antiviral activity.Autophagy. 2023 Aug;19(8):2257-2274. doi: 10.1080/15548627.2023.2179844. Epub 2023 Feb 27. Autophagy. 2023. PMID: 36779599 Free PMC article.
-
Biochemical evidence that the whole compartment activity behavior of GAPDH differs between the cytoplasm and nucleus.PLoS One. 2023 Aug 31;18(8):e0290892. doi: 10.1371/journal.pone.0290892. eCollection 2023. PLoS One. 2023. PMID: 37651389 Free PMC article.
-
Structural basis of catalytic activation in human splicing.Nature. 2023 May;617(7962):842-850. doi: 10.1038/s41586-023-06049-w. Epub 2023 May 10. Nature. 2023. PMID: 37165190 Free PMC article.
-
Spliceosomal helicases DDX41/SACY-1 and PRP22/MOG-5 both contribute to proofreading against proximal 3' splice site usage.RNA. 2024 Mar 18;30(4):404-417. doi: 10.1261/rna.079888.123. RNA. 2024. PMID: 38282418 Free PMC article.
-
Unveiling the DHX15-G-patch interplay in retroviral RNA packaging.Proc Natl Acad Sci U S A. 2024 Oct;121(40):e2407990121. doi: 10.1073/pnas.2407990121. Epub 2024 Sep 25. Proc Natl Acad Sci U S A. 2024. PMID: 39320912 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

