Structural intermediates in the low pH-induced transition of influenza hemagglutinin
- PMID: 33253316
- PMCID: PMC7728236
- DOI: 10.1371/journal.ppat.1009062
Structural intermediates in the low pH-induced transition of influenza hemagglutinin
Abstract
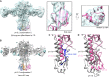
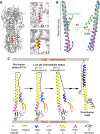
The hemagglutinin (HA) glycoproteins of influenza viruses play a key role in binding host cell receptors and in mediating virus-host cell membrane fusion during virus infection. Upon virus entry, HA is triggered by low pH and undergoes large structural rearrangements from a prefusion state to a postfusion state. While structures of prefusion state and postfusion state of HA have been reported, the intermediate structures remain elusive. Here, we report two distinct low pH intermediate conformations of the influenza virus HA using cryo-electron microscopy (cryo-EM). Our results show that a decrease in pH from 7.8 to 5.2 triggers the release of fusion peptides from the binding pockets and then causes a dramatic conformational change in the central helices, in which the membrane-proximal ends of the central helices unwind to an extended form. Accompanying the conformational changes of the central helices, the stem region of the HA undergoes an anticlockwise rotation of 9.5 degrees and a shift of 15 Å. The HA head, after being stabilized by an antibody, remains unchanged compared to the neutral pH state. Thus, the conformational change of the HA stem region observed in our research is likely to be independent of the HA head. These results provide new insights into the structural transition of HA during virus entry.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Reversible structural changes in the influenza hemagglutinin precursor at membrane fusion pH.Proc Natl Acad Sci U S A. 2022 Aug 16;119(33):e2208011119. doi: 10.1073/pnas.2208011119. Epub 2022 Aug 8. Proc Natl Acad Sci U S A. 2022. PMID: 35939703 Free PMC article.
-
Structural characterization of an early fusion intermediate of influenza virus hemagglutinin.J Virol. 2011 May;85(10):5172-82. doi: 10.1128/JVI.02430-10. Epub 2011 Mar 2. J Virol. 2011. PMID: 21367895 Free PMC article.
-
Probing the metastable state of influenza hemagglutinin.J Biol Chem. 2017 Dec 29;292(52):21590-21597. doi: 10.1074/jbc.M117.815043. Epub 2017 Nov 10. J Biol Chem. 2017. PMID: 29127198 Free PMC article.
-
Influenza virus entry.Adv Exp Med Biol. 2012;726:201-21. doi: 10.1007/978-1-4614-0980-9_9. Adv Exp Med Biol. 2012. PMID: 22297515 Free PMC article. Review.
-
Influenza virus-mediated membrane fusion: Structural insights from electron microscopy.Arch Biochem Biophys. 2015 Sep 1;581:86-97. doi: 10.1016/j.abb.2015.04.011. Epub 2015 May 6. Arch Biochem Biophys. 2015. PMID: 25958107 Free PMC article. Review.
Cited by
-
Engineering a cleaved, prefusion-stabilized influenza B virus hemagglutinin by identification and locking of all six pH switches.PNAS Nexus. 2024 Oct 11;3(10):pgae462. doi: 10.1093/pnasnexus/pgae462. eCollection 2024 Oct. PNAS Nexus. 2024. PMID: 39445049 Free PMC article.
-
Molecular Dynamics Investigation of the Influenza Hemagglutinin Conformational Changes in Acidic pH.J Phys Chem B. 2024 Nov 14;128(45):11151-11163. doi: 10.1021/acs.jpcb.4c04607. Epub 2024 Nov 4. J Phys Chem B. 2024. PMID: 39497238 Free PMC article.
-
Structural and pKa Estimation of the Amphipathic HR1 in SARS-CoV-2: Insights from Constant pH MD, Linear vs. Nonlinear Normal Mode Analysis.Int J Mol Sci. 2023 Nov 10;24(22):16190. doi: 10.3390/ijms242216190. Int J Mol Sci. 2023. PMID: 38003380 Free PMC article.
-
Characterization of Neutralizing Monoclonal Antibodies and Identification of a Novel Conserved C-Terminal Linear Epitope on the Hemagglutinin Protein of the H9N2 Avian Influenza Virus.Viruses. 2022 Nov 15;14(11):2530. doi: 10.3390/v14112530. Viruses. 2022. PMID: 36423139 Free PMC article.
-
Reversible structural changes in the influenza hemagglutinin precursor at membrane fusion pH.Proc Natl Acad Sci U S A. 2022 Aug 16;119(33):e2208011119. doi: 10.1073/pnas.2208011119. Epub 2022 Aug 8. Proc Natl Acad Sci U S A. 2022. PMID: 35939703 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

